2IY3
 
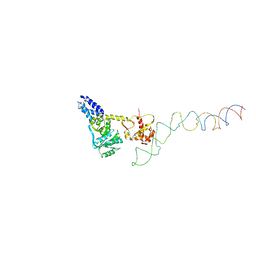 | | Structure of the E. Coli Signal Regognition Particle | | Descriptor: | 4.5S RNA, SIGNAL SEQUENCE, Signal recognition particle protein,Signal recognition particle 54 kDa protein | | Authors: | Schaffitzel, C, Oswald, M, Berger, I, Ishikawa, T, Abrahams, J.P, Koerten, H.K, Koning, R.I, Ban, N. | | Deposit date: | 2006-07-12 | | Release date: | 2006-11-02 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structure of the E. Coli Signal Recognition Particle Bound to a Translating Ribosome
Nature, 444, 2006
|
|
5MG3
 
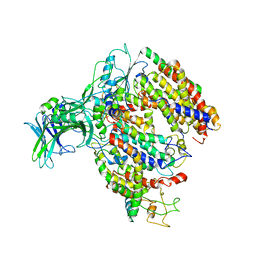 | | EM fitted model of bacterial holo-translocon | | Descriptor: | Membrane protein insertase YidC, Protein translocase subunit SecD, Protein translocase subunit SecE, ... | | Authors: | Schaffitzel, C, Botte, M. | | Deposit date: | 2016-11-20 | | Release date: | 2016-12-28 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion.
Sci Rep, 6, 2016
|
|
6FEC
 
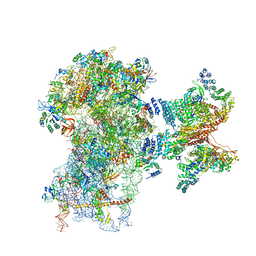 | | Human cap-dependent 48S pre-initiation complex | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schaffitzel, C, Schaffitzel, C. | | Deposit date: | 2017-12-31 | | Release date: | 2018-03-14 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Structure of a human cap-dependent 48S translation pre-initiation complex.
Nucleic Acids Res., 46, 2018
|
|
8COI
 
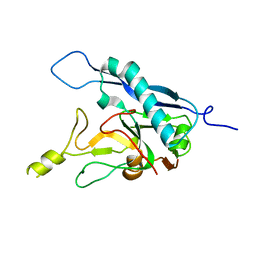 | | Human adenovirus-derived synthetic ADDobody binder | | Descriptor: | ADDobody | | Authors: | Buzas, D, Toelzer, C, Gupta, K, Berger-Schaffitzel, C, Berger, I. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
6HCR
 
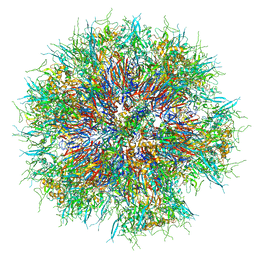 | | Synthetic Self-assembling ADDomer Platform for Highly Efficient Vaccination by Genetically-encoded Multi-epitope Display | | Descriptor: | Penton protein | | Authors: | Bufton, J.C, Berger, I, Schaffitzel, C, Garzoni, F, Rabi, F.A. | | Deposit date: | 2018-08-16 | | Release date: | 2019-08-28 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Synthetic self-assembling ADDomer platform for highly efficient vaccination by genetically encoded multiepitope display.
Sci Adv, 5, 2019
|
|
7QG6
 
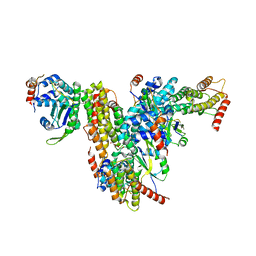 | | Co-crystal structure of UPF3A-RRM-NOPS-L with UPF2-MIF4GIII | | Descriptor: | CHLORIDE ION, Regulator of nonsense transcripts 2, Regulator of nonsense transcripts 3A, ... | | Authors: | Powers, K.T, Bufton, J.C, Szeto, J.A, Schaffitzel, C. | | Deposit date: | 2021-12-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of nonsense-mediated mRNA decay factors UPF3B and UPF3A in complex with UPF2 reveal molecular basis for competitive binding and for neurodevelopmental disorder-causing mutation.
Nucleic Acids Res., 50, 2022
|
|
8F2U
 
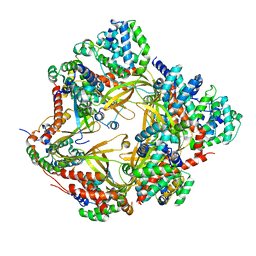 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8F2R
 
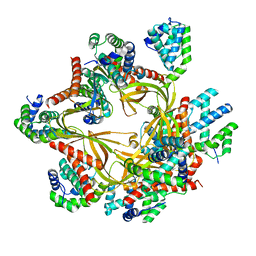 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
6EMK
 
 | | Cryo-EM Structure of Saccharomyces cerevisiae Target of Rapamycin Complex 2 | | Descriptor: | Serine/threonine-protein kinase TOR2, Target of rapamycin complex 2 subunit AVO1, Target of rapamycin complex 2 subunit AVO2, ... | | Authors: | Karuppasamy, M, Kusmider, B, Oliveira, T.M, Gaubitz, C, Prouteau, M, Loewith, R, Schaffitzel, C. | | Deposit date: | 2017-10-02 | | Release date: | 2017-12-06 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Cryo-EM structure of Saccharomyces cerevisiae target of rapamycin complex 2.
Nat Commun, 8, 2017
|
|
4V5B
 
 | | Structure of PDF binding helix in complex with the ribosome. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Bingel-Erlenmeyer, R, Kohler, R, Kramer, G, Sandikci, A, Antolic, S, Maier, T, Schaffitzel, C, Wiedmann, B, Bukau, B, Ban, N. | | Deposit date: | 2007-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | A Peptide Deformylase-Ribosome Complex Reveals Mechanism of Nascent Chain Processing.
Nature, 452, 2008
|
|
7ZH2
 
 | | SARS CoV Spike protein, Closed C1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH5
 
 | | SARS CoV Spike protein, Open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH1
 
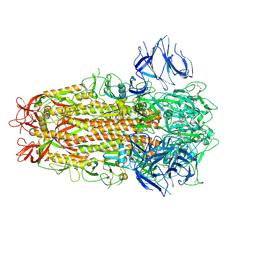 | | SARS CoV Spike protein, Closed C3 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
6ZB5
 
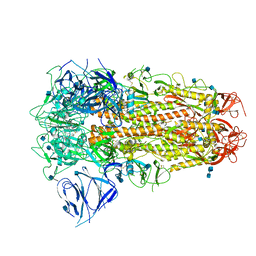 | | SARS CoV-2 Spike protein, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
6ZB4
 
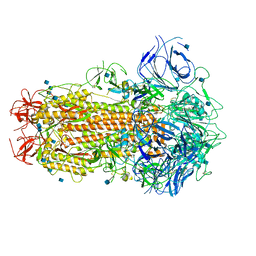 | | SARS CoV-2 Spike protein, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
8QB3
 
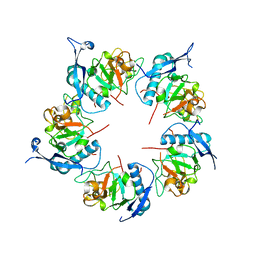 | | ADDobody zinc containing condition | | Descriptor: | ADDobody, ZINC ION | | Authors: | Buzas, D, Toelzer, C, Berger, I, Schaffitzel, C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
7NWU
 
 | | Co-crystal structure of UPF3B-RRM-NOPS-L with UPF2-MIF4GIII | | Descriptor: | PENTAETHYLENE GLYCOL, Regulator of nonsense transcripts 2, Regulator of nonsense transcripts 3B, ... | | Authors: | Powers, K.T, Bufton, J.C, Szeto, J.A, Schaffitzel, C. | | Deposit date: | 2021-03-17 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of nonsense-mediated mRNA decay factors UPF3B and UPF3A in complex with UPF2 reveal molecular basis for competitive binding and for neurodevelopmental disorder-causing mutation.
Nucleic Acids Res., 50, 2022
|
|
8C9N
 
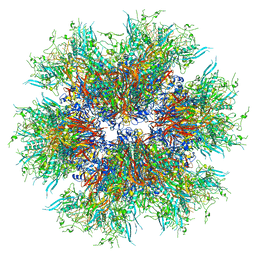 | | MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle | | Descriptor: | Penton protein | | Authors: | Bufton, J.C, Capin, J, Boruku, U, Garzoni, F, Schaffitzel, C, Berger, I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | In vitro generated antibodies guide thermostable ADDomer nanoparticle design for nasal vaccination and passive immunization against SARS-CoV-2.
Antib Ther, 6, 2023
|
|
5AKA
 
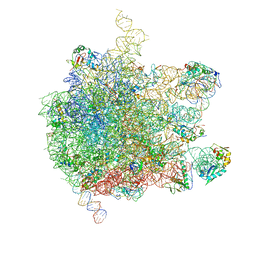 | | EM structure of ribosome-SRP-FtsY complex in closed state | | Descriptor: | 23S ribosomal RNA, 4.5S ribosomal RNA, 50S RIBOSOMAL PROTEIN L11, ... | | Authors: | von Loeffelholz, O, Jiang, Q, Ariosa, A, Karuppasamy, M, Huard, K, Berger, I, Shan, S, Schaffitzel, C. | | Deposit date: | 2015-03-03 | | Release date: | 2015-03-25 | | Last modified: | 2017-12-20 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Ribosome-Srp-Ftsy Cotranslational Targeting Complex in the Closed State.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2AKI
 
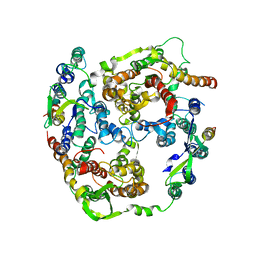 | | Normal mode-based flexible fitted coordinates of a translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
2AKH
 
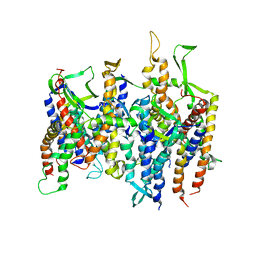 | | Normal mode-based flexible fitted coordinates of a non-translocating SecYEG protein-conducting channel into the cryo-EM map of a SecYEG-nascent chain-70S ribosome complex from E. coli | | Descriptor: | Preprotein translocase secE subunit, Preprotein translocase secY subunit, Protein-export membrane protein secG | | Authors: | Mitra, K.M, Schaffitzel, C, Shaikh, T, Tama, F, Jenni, S, Brooks III, C.L, Ban, N, Frank, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-11-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.9 Å) | | Cite: | Structure of the E. coli protein-conducting channel bound to a translating ribosome.
Nature, 438, 2005
|
|
2VRH
 
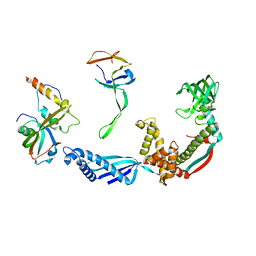 | | Structure of the E. coli trigger factor bound to a translating ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, 50S RIBOSOMAL PROTEIN L29, ... | | Authors: | Merz, F, Boehringer, D, Schaffitzel, C, Preissler, S, Hoffmann, A, Maier, T, Rutkowska, A, Lozza, J, Ban, N, Bukau, B, Deuerling, E. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-17 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Molecular Mechanism and Structure of Trigger Factor Bound to the Translating Ribosome.
Embo J., 27, 2008
|
|
2XKV
 
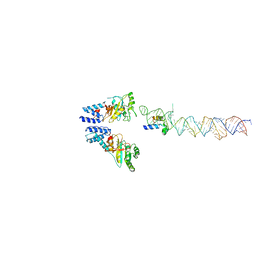 | | Atomic Model of the SRP-FtsY Early Conformation | | Descriptor: | 4.5S RNA, CELL DIVISION PROTEIN FTSY, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Estrozi, L.F, Boehringer, D, Shan, S.-o, Ban, N, Schaffitzel, C. | | Deposit date: | 2010-07-13 | | Release date: | 2010-12-15 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Cryo-Em Structure of the E. Coli Translating Ribosome in Complex with Srp and its Receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7ODL
 
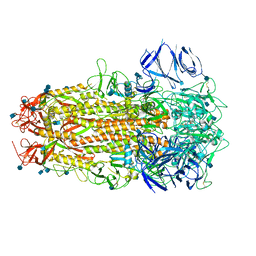 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7OD3
 
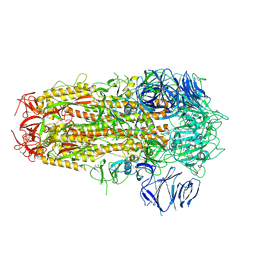 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-28 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
