1HOY
 
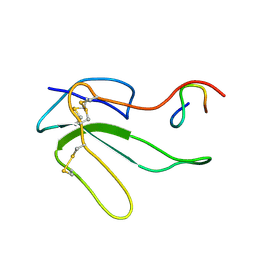 | | NMR STRUCTURE OF THE COMPLEX BETWEEN A-BUNGAROTOXIN AND A MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Niccolai, N, Neri, P. | | Deposit date: | 2000-12-12 | | Release date: | 2000-12-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1JBD
 
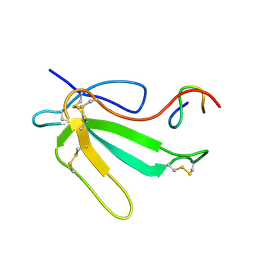 | | NMR Structure of the Complex Between alpha-bungarotoxin and a Mimotope of the Nicotinic Acetylcholine Receptor | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Klein, S, Niccolai, N. | | Deposit date: | 2001-06-04 | | Release date: | 2001-06-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
2KC0
 
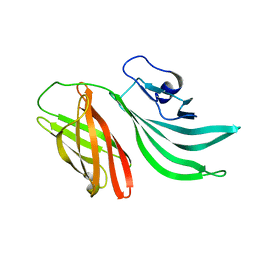 | | Solution structure of the factor H binding protein | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Veggi, D, Dragonetti, S, Savino, S, Scarselli, M, Romagnoli, G, Pizza, M, Banci, L, Rappuoli, R. | | Deposit date: | 2008-12-13 | | Release date: | 2009-02-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Factor H-binding Protein, a Survival Factor and Protective Antigen of Neisseria meningitidis
J.Biol.Chem., 284, 2009
|
|
1YS5
 
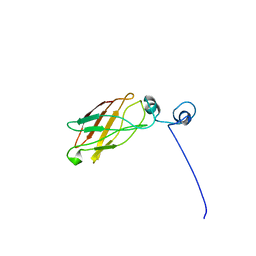 | | Solution structure of the antigenic domain of GNA1870 of Neisseria meningitidis | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Savino, S, Masignani, V, Pizza, M, Scarselli, M, Swennen, E, Romagnoli, G, Veggi, D, Banci, L, Rappuoli, R. | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis
J.Biol.Chem., 281, 2006
|
|
4DMW
 
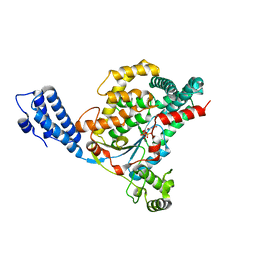 | | Crystal structure of the GT domain of Clostridium difficile toxin A (TcdA) in complex with UDP and Manganese | | Descriptor: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Scarselli, M, Maione, D, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
4DMV
 
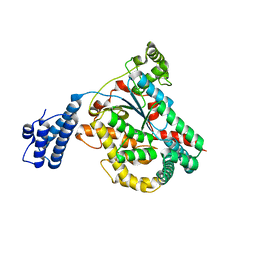 | | Crystal structure of the GT domain of Clostridium difficile Toxin A | | Descriptor: | Toxin A | | Authors: | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Maione, D, Scarselli, M, Martinelli, M. | | Deposit date: | 2012-02-08 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
2L4O
 
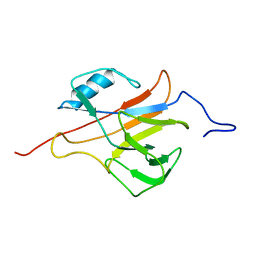 | | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain | | Descriptor: | Cell wall surface anchor family protein | | Authors: | Gentile, M.A, Melchiorre, S, Emolo, C, Moschioni, M, Gianfaldoni, C, Pancotto, L, Ferlenghi, I, Scarselli, M, Pansegrau, W, Veggi, D, Merola, M, Cantini, F, Ruggiero, P, Banci, L, Masignani, V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-03-02 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain: in vivo protection and implications for the molecular mechanisms of pilus polymerization
To be Published
|
|
2N6J
 
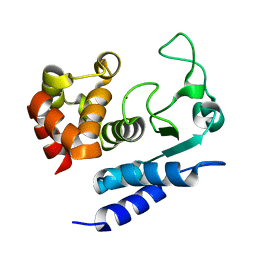 | | Solution structure of Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile | | Descriptor: | ZINC ION, Zinc metalloprotease Zmp1 | | Authors: | Banci, L, Cantini, F, Scarselli, M, Rubino, J.T, Martinelli, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-01-13 | | Last modified: | 2016-04-06 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of zinc-bound Zmp1, a zinc-dependent metalloprotease secreted by Clostridium difficile.
J.Biol.Inorg.Chem., 21, 2016
|
|
1P83
 
 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | Descriptor: | 10 kDa chaperonin | | Authors: | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | Deposit date: | 2003-05-06 | | Release date: | 2003-05-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
1P82
 
 | | NMR STRUCTURE OF 1-25 FRAGMENT OF MYCOBACTERIUM TUBERCULOSIS CPN10 | | Descriptor: | 10 kDa chaperonin | | Authors: | Ciutti, A, Spiga, O, Giannozzi, E, Scarselli, M, Di Maro, D, Calamandrei, D, Niccolai, N, Bernini, A. | | Deposit date: | 2003-05-06 | | Release date: | 2003-05-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 1-25 fragment of Cpn10 from Mycobacterium Tuberculosis
To be Published
|
|
2X9W
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, GLYCEROL | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9Z
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9X
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, IMIDAZOLE, SODIUM ION | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9Y
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
1RGJ
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN ALPHA-BUNGAROTOXIN AND MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR WITH ENHANCED ACTIVITY | | Descriptor: | MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR, long neurotoxin 1 | | Authors: | Bernini, A, Spiga, O, Ciutti, A, Scarselli, M, Bracci, L, Lozzi, L, Lelli, B, Neri, P, Niccolai, N. | | Deposit date: | 2003-11-12 | | Release date: | 2003-11-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR and MD studies on the interaction between ligand peptides and alpha-bungarotoxin.
J.Mol.Biol., 339, 2004
|
|
1IK8
 
 | | NMR structure of Alpha-Bungarotoxin | | Descriptor: | LONG NEUROTOXIN 1 | | Authors: | Niccolai, N, Ciutti, A, Spiga, O. | | Deposit date: | 2001-05-03 | | Release date: | 2001-05-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1IKC
 
 | | NMR Structure of alpha-Bungarotoxin | | Descriptor: | long neurotoxin 1 | | Authors: | Niccolai, N, Spiga, O, Ciutti, A. | | Deposit date: | 2001-05-03 | | Release date: | 2001-05-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
2Y7S
 
 | |
5T5F
 
 | |
4CJD
 
 | | Crystal structure of Neisseria meningitidis trimeric autotransporter and vaccine antigen NadA | | Descriptor: | IODIDE ION, NADA | | Authors: | Malito, E, Biancucci, M, Spraggon, G, Bottomley, M.J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-26 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Structure of the Meningococcal Vaccine Antigen Nada and Epitope Mapping of a Bactericidal Antibody.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2LFU
 
 | | The structure of a N. meningitides protein targeted for vaccine development | | Descriptor: | Gna2132 | | Authors: | Esposito, V, Musi, V, De Chiara, C, Kelly, G, Veggi, D, Pizza, M, Pastore, A. | | Deposit date: | 2011-07-15 | | Release date: | 2011-10-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal Domain of Neisseria Heparin Binding Antigen (NHBA), One of the Main Antigens of a Novel Vaccine against Neisseria meningitidis.
J.Biol.Chem., 286, 2011
|
|
