5N9X
 
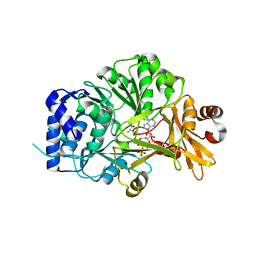 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, ligand bound structure | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylation domain, MAGNESIUM ION, ... | | Authors: | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
5N9W
 
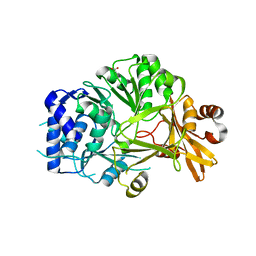 | | Structure of adenylation domain THR1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces SP.OH-5093, apo structure | | Descriptor: | ACETATE ION, Adenylation domain | | Authors: | Savino, C, Vallone, B, Scaglione, A, Parisi, G, Montemiglio, L.C, Fullone, M.R, Grgurina, I. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.456 Å) | | Cite: | Structure of the adenylation domain Thr1 involved in the biosynthesis of 4-chlorothreonine in Streptomyces sp. OH-5093-protein flexibility and molecular bases of substrate specificity.
FEBS J., 284, 2017
|
|
4XE3
 
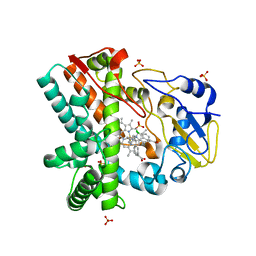 | | OleP, the cytochrome P450 epoxidase from Streptomyces antibioticus involved in Oleandomycin biosynthesis: functional analysis and crystallographic structure in complex with clotrimazole. | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Montemiglio, L.C, Parisi, G, Scaglione, A, Savino, C, Vallone, B. | | Deposit date: | 2014-12-22 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional analysis and crystallographic structure of clotrimazole bound OleP, a cytochrome P450 epoxidase from Streptomyces antibioticus involved in oleandomycin biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
6H6J
 
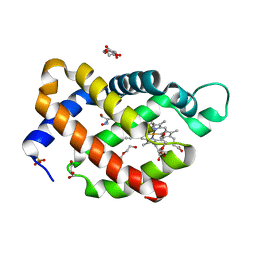 | | Carbomonoxy murine neuroglobin Gly-loop mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBON MONOXIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H5Z
 
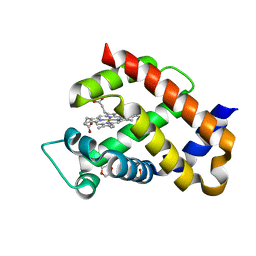 | | Ferric murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-25 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H6I
 
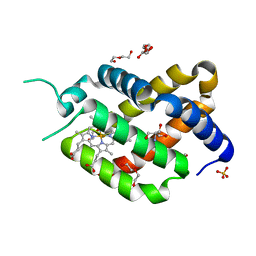 | | Ferric murine neuroglobin Gly-loop mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
6H6C
 
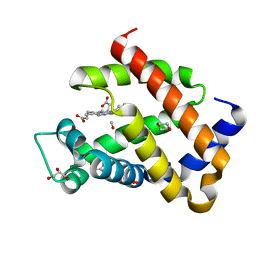 | | Carbomonoxy murine neuroglobin F106A mutant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CARBON MONOXIDE, GLYCEROL, ... | | Authors: | Exertier, C, Vallone, B, Savino, C, Freda, I, Montemiglio, L.C, Cerutti, G, Scaglione, A, Parisi, G. | | Deposit date: | 2018-07-27 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Proximal and distal control for ligand binding in neuroglobin: role of the CD loop and evidence for His64 gating.
Sci Rep, 9, 2019
|
|
5MNS
 
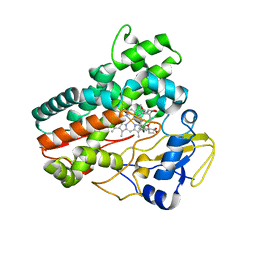 | | Structural and functional characterization of OleP in complex with 6DEB in sodium formate | | Descriptor: | 6-DEOXYERYTHRONOLIDE B, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Parisi, G, Savino, C, Montemiglio, L.C, Vallone, B. | | Deposit date: | 2016-12-13 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Substrate-induced conformational change in cytochrome P450 OleP.
FASEB J., 33, 2019
|
|
5MNV
 
 | |
5BZB
 
 | | NavMs voltage-gated sodium channel pore and C-terminal domain | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2015-06-11 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4X8A
 
 | | NavMS pore and C-terminal domain grown from protein purified in LiCl | | Descriptor: | HEGA-10, Ion transport protein, NONAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4X88
 
 | | E178D Selectivity filter mutant of NavMS voltage-gated pore | | Descriptor: | HEGA-10, Ion transport protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4X89
 
 | | NavMs voltage-gated sodium channal pore and C-terminal domain soaked with Silver nitrate | | Descriptor: | HEGA-10, Ion transport protein, PENTAETHYLENE GLYCOL, ... | | Authors: | Naylor, C.E, Bagneris, C, Wallace, B.A. | | Deposit date: | 2014-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Molecular basis of ion permeability in a voltage-gated sodium channel.
Embo J., 35, 2016
|
|
4MU5
 
 | | Crystal structure of murine neuroglobin mutant M144W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vallone, B, Avella, G, Savino, C, Ardiccioni, C, Brunori, M. | | Deposit date: | 2013-09-20 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O1T
 
 | | Crystal structure of murine neuroglobin mutant F106W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-16 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O2G
 
 | | Crystal structure of carbomonoxy murine neuroglobin mutant V140W | | Descriptor: | CARBON MONOXIDE, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-17 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4NZI
 
 | | Crystal structure of murine neuroglobin mutant V140W | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-12 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O35
 
 | | Crystal structure of carbomonoxy murine neuroglobin mutant F106W | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CARBON MONOXIDE, Neuroglobin, ... | | Authors: | Avella, G, Savino, C, Vallone, B. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the internal cavity of neuroglobin demonstrates the role of the haem-sliding mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
