1KR4
 
 | | Structure Genomics, Protein TM1056, cutA | | Descriptor: | Protein TM1056, cutA | | Authors: | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
1P9Q
 
 | | Structure of a hypothetical protein AF0491 from Archaeoglobus fulgidus | | Descriptor: | Hypothetical protein AF0491 | | Authors: | Savchenko, A, Evdokimova, E, Skarina, T, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A. | | Deposit date: | 2003-05-12 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism.
J.Biol.Chem., 280, 2005
|
|
1QW2
 
 | | Crystal Structure of a Protein of Unknown Function TA1206 from Thermoplasma acidophilum | | Descriptor: | conserved hypothetical protein TA1206 | | Authors: | Savchenko, A, Evdokimova, E, Kudrytska, M, Edwards, A.E, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-30 | | Release date: | 2004-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of a Hypothetical Protein "TA1206" from Thermoplasma acidophilum
To be Published
|
|
4W8K
 
 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
1K7K
 
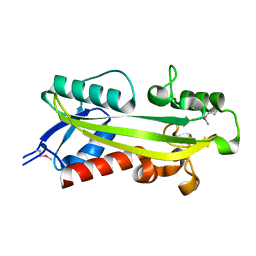 | | crystal structure of RdgB- inosine triphosphate pyrophosphatase from E. coli | | Descriptor: | Hypothetical protein yggV | | Authors: | Sanishvili, R, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-19 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
2PYU
 
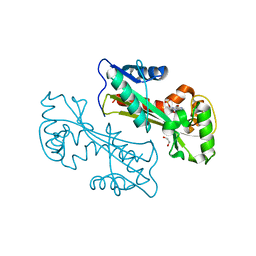 | | Structure of the E. coli inosine triphosphate pyrophosphatase RgdB in complex with IMP | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, Inosine Triphosphate Pyrophosphatase RdgB | | Authors: | Singer, A.U, Proudfoot, M, Skarina, T, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2007-05-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
2Q16
 
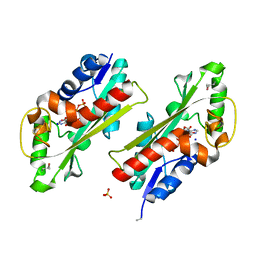 | | Structure of the E. coli inosine triphosphate pyrophosphatase RgdB in complex with ITP | | Descriptor: | CALCIUM ION, HAM1 protein homolog, INOSINE 5'-TRIPHOSPHATE, ... | | Authors: | Singer, A.U, Lam, R, Proudfoot, M, Skarina, T, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2007-05-23 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
6U69
 
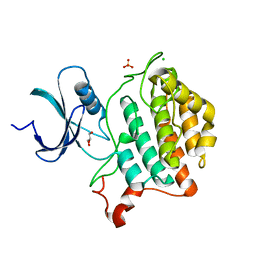 | | Crystal structure of Yck2 from Candida albicans, apoenzyme | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Overcoming Fungal Echinocandin Resistance through Inhibition of the Non-essential Stress Kinase Yck2.
Cell Chem Biol, 27, 2020
|
|
8SDC
 
 | | Crystal structure of fluoroacetate dehalogenase Daro3835 apoenzyme | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Iakounine, A, Savchenko, A. | | Deposit date: | 2023-04-06 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases.
Febs J., 290, 2023
|
|
8SDD
 
 | | Crystal structure of fluoroacetate dehalogenase Daro3835 H274N mutant with D107-glycolyl intermediate | | Descriptor: | Alpha/beta hydrolase fold protein | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Iakounine, A, Savchenko, A. | | Deposit date: | 2023-04-06 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases.
Febs J., 290, 2023
|
|
8SCD
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with reaction intermediate | | Descriptor: | 2-amino-6-methylidene-6,7-dihydropteridin-4(3H)-one, 4-AMINOBENZOIC ACID, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-05 | | Release date: | 2023-05-03 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
4W8I
 
 | | Crystal structure of LpSPL/Lpp2128, Legionella pneumophila sphingosine-1 phosphate lyase | | Descriptor: | Probable sphingosine-1-phosphate lyase | | Authors: | Stogios, P.J, Daniels, C, Skarina, T, Cuff, M, Di Leo, R, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4W97
 
 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
4WZ2
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | Authors: | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-28 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
8DVC
 
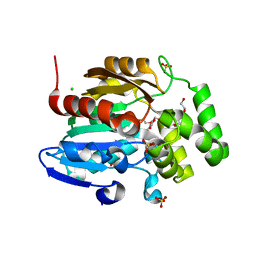 | | Receptor ShHTL5 from Striga hermonthica in complex with strigolactone agonist GR24 | | Descriptor: | (3R,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methyl)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Arellano-Saab, A, Skarina, T, Yim, V, Savchenko, A, Stogios, P.J, McCourt, P. | | Deposit date: | 2022-07-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.638 Å) | | Cite: | Structural analysis of a hormone-bound Striga strigolactone receptor.
Nat.Plants, 9, 2023
|
|
7MH7
 
 | |
5SUJ
 
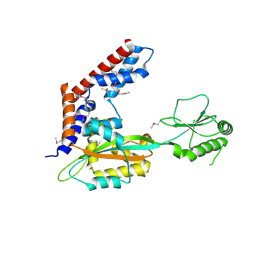 | | Crystal structure of uncharacterized protein LPG2148 from Legionella pneumophila | | Descriptor: | Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.356 Å) | | Cite: | Discovery of Ubiquitin Deamidases in the Pathogenic Arsenal of Legionella pneumophila.
Cell Rep, 23, 2018
|
|
6XS4
 
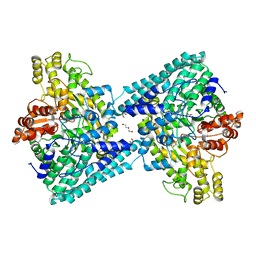 | | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Formate C-acetyltransferase | | Authors: | Valleau, D, Evdokimova, E, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae.
To Be Published
|
|
4WQK
 
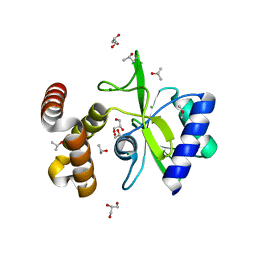 | | Crystal structure of aminoglycoside nucleotidylyltransferase ANT(2")-Ia, apo form | | Descriptor: | 2''-aminoglycoside nucleotidyltransferase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cox, G, Stogios, P.J, Savchenko, A, Wright, G.D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural and Molecular Basis for Resistance to Aminoglycoside Antibiotics by the Adenylyltransferase ANT(2)-Ia.
Mbio, 6, 2015
|
|
4WJ0
 
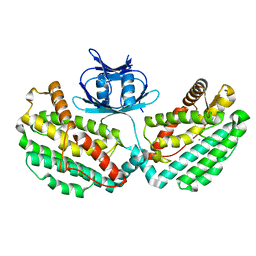 | | Structure of PH1245, a cas1 from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, CRISPR-associated endonuclease Cas1 | | Authors: | Petit, P, Brown, G, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of PH1245, a cas1 from Pyrococcus horikoshii
To Be Published
|
|
8EJV
 
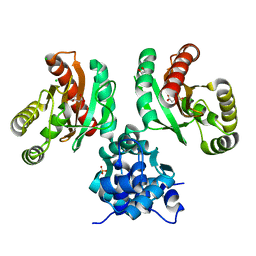 | | The crystal structure of Pseudomonas putida PcaR in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR in complex with succinate
To Be Published
|
|
8EJU
 
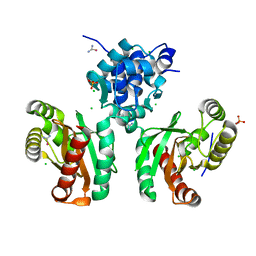 | | The crystal structure of Pseudomonas putida PcaR | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Transcription regulatory protein (Pca regulon), ... | | Authors: | Pham, C, Skarina, T, Di Leo, R, Stogios, P.J, Mahadevan, R, Savchenko, A. | | Deposit date: | 2022-09-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The crystal structure of Pseudomonas putida PcaR
To Be Published
|
|
7SNU
 
 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | Descriptor: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
4E0B
 
 | | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6 | | Descriptor: | ACETATE ION, Malate dehydrogenase | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-02 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | 2.17 Angstrom resolution crystal structure of malate dehydrogenase from Vibrio vulnificus CMCP6
To be Published
|
|
4DCA
 
 | | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, ADP-bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside phosphotransferase, MAGNESIUM ION | | Authors: | Stogios, P.J, Minasov, G, Singer, A.U, Tan, K, Nocek, B, Evdokimova, E, Egorova, O, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-01-17 | | Release date: | 2012-02-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aminoglycoside phosphotransferase APH(2'')-Ib, ADP-bound
TO BE PUBLISHED
|
|
