6ZVA
 
 | | Crystal structure of My5 | | Descriptor: | Myomesin-1, SULFATE ION | | Authors: | Sauer, F, Wilmanns, M. | | Deposit date: | 2020-07-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of Myomesin-1
To Be Published
|
|
3KNB
 
 | |
3PUC
 
 | |
3Q5O
 
 | |
3QP3
 
 | |
6TUN
 
 | | Helicase domain complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, CHLORIDE ION, General transcription and DNA repair factor IIH helicase subunit XPD | | Authors: | Sauer, F, Kisker, C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | In TFIIH the Arch domain of XPD is mechanistically essential for transcription and DNA repair.
Nat Commun, 11, 2020
|
|
4F14
 
 | |
6H4L
 
 | | Structure of Titin M4 trigonal form | | Descriptor: | CHLORIDE ION, Titin, ZINC ION | | Authors: | Sauer, F, Wilmanns, M. | | Deposit date: | 2018-07-21 | | Release date: | 2019-08-07 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural diversity in the atomic resolution 3D fingerprint of the titin M-band segment.
Plos One, 14, 2019
|
|
6H4J
 
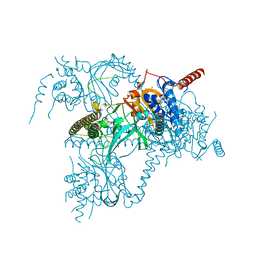 | | Usp25 catalytic domain | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4I
 
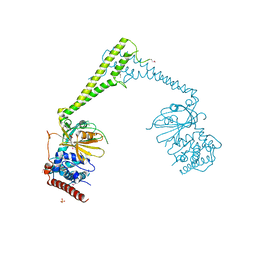 | | Usp28 catalytic domain apo | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4H
 
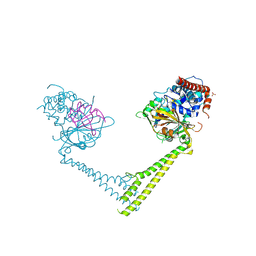 | | Usp28 catalytic domain variant E593D in complex with UbPA | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28, ... | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4K
 
 | | Structure of the Usp25 C-terminal domain | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
5B5Q
 
 | |
5LST
 
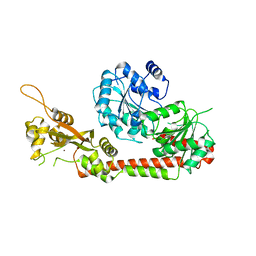 | | Crystal structure of the human RecQL4 helicase. | | Descriptor: | ATP-dependent DNA helicase Q4, ZINC ION | | Authors: | Kaiser, S, Sauer, F, Kisker, C. | | Deposit date: | 2016-09-05 | | Release date: | 2017-07-05 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The structural and functional characterization of human RecQ4 reveals insights into its helicase mechanism.
Nat Commun, 8, 2017
|
|
6HCI
 
 | |
6FDU
 
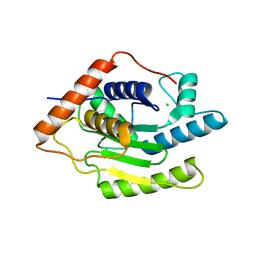 | | Structure of Chlamydia trachomatis effector protein Cdu1 bound to Compound 3 | | Descriptor: | (2~{S},3~{S})-2-[[(2~{S})-2-[3,5-bis(chloranyl)phenyl]-2-(dimethylamino)ethanoyl]amino]-~{N}-[[2-(iminomethyl)pyrimidin-4-yl]methyl]-3-methyl-pentanamide, CHLORIDE ION, Deubiquitinase and deneddylase Dub1 | | Authors: | Ramirez, Y, Kisker, C, Altmann, E. | | Deposit date: | 2017-12-26 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Substrate Recognition and Covalent Inhibition of Cdu1 from Chlamydia trachomatis.
ChemMedChem, 13, 2018
|
|
6FDQ
 
 | |
6FDK
 
 | |
