2D7H
 
 | | Crystal structure of the ccc complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*CP*CP*C)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7G
 
 | | Crystal structure of the aa complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7E
 
 | | Crystal structure of N-terminal domain of PriA from E.coli | | Descriptor: | Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-18 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWN
 
 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | DNA (5'-D(*A*G)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWL
 
 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*(DC))-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2DWM
 
 | | Crystal structure of the PriA protein complexed with oligonucleotides | | Descriptor: | 5'-D(*AP*T)-3', Primosomal protein N | | Authors: | Sasaki, K, Ose, T, Tanaka, T, Masai, H, Maenaka, K, Kohda, D. | | Deposit date: | 2006-08-15 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
1GCN
 
 | |
3W3D
 
 | | Crystal structure of smooth muscle G actin DNase I complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, gamma-enteric smooth muscle, ... | | Authors: | Sakabe, N, Sakabe, K, Sasaki, K, Kondo, H, Shimomur, M. | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Refined structure and solvent network of chicken gizzard G-actin DNase 1 complex at 1.8A resolution
Acta Crystallogr.,Sect.A, 49, 1993
|
|
5AXV
 
 | | Crystal Structure of Cypovirus Polyhedra R13K Mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
5AXU
 
 | | Crystal Structure of Cypovirus Polyhedra R13A Mutant | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Abe, S, Ijiri, H, Negishi, H, Yamanaka, H, Sasaki, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2015-08-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Enzyme-Encapsulated Protein Containers by In Vivo Crystal Engineering
Adv. Mater. Weinheim, 27, 2015
|
|
3A8U
 
 | |
2ZB6
 
 | | Crystal structure of the measles virus hemagglutinin (oligo-sugar type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin protein | | Authors: | Hashiguchi, T, Kajikawa, M, Maita, N, Takeda, M, Kuroki, K, Sasaki, K, Kohda, D, Yanagi, Y, Maenaka, K. | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of measles virus hemagglutinin provides insight into effective vaccines
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3W7Y
 
 | |
3W80
 
 | |
2ZB5
 
 | | Crystal structure of the measles virus hemagglutinin (complex-sugar-type) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin protein | | Authors: | Hashiguchi, T, Kajikawa, M, Maita, N, Takeda, M, Kuroki, K, Sasaki, K, Kohda, D, Yanagi, Y, Maenaka, K. | | Deposit date: | 2007-10-16 | | Release date: | 2007-11-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of measles virus hemagglutinin provides insight into effective vaccines
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3W7Z
 
 | |
1UGL
 
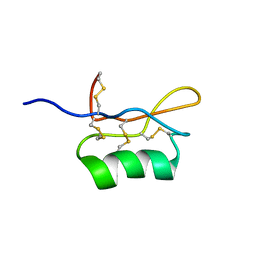 | | Solution structure of S8-SP11 | | Descriptor: | S-locus pollen protein | | Authors: | Mishima, M, Takayama, S, Sasaki, K, Jee, J.G, Kojima, C, Isogai, A, Shirakawa, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the Male Determinant Factor for Brassica Self-incompatibility
J.Biol.Chem., 278, 2003
|
|
3W3E
 
 | |
8GS6
 
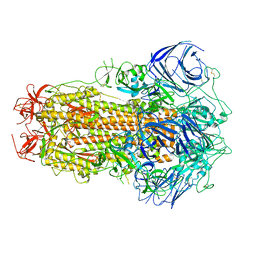 | | Structure of the SARS-CoV-2 BA.2.75 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Tabata-Sasaki, K, Kita, S, Fukuhara, H, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2022-09-05 | | Release date: | 2022-10-26 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant.
Cell Host Microbe, 30, 2022
|
|
3GOI
 
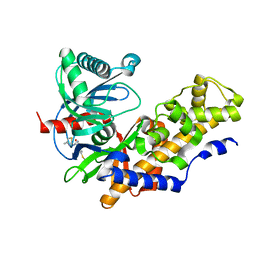 | | Human glucokinase in complex with a synthetic activator | | Descriptor: | 2-(methylamino)-N-(4-methyl-1,3-thiazol-2-yl)-5-[(4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl]benzamide, Glucokinase, alpha-D-glucopyranose | | Authors: | Kamata, K, Mitsuya, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Discovery of novel 3,6-disubstituted 2-pyridinecarboxamide derivatives as GK activators
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3H1V
 
 | | Human glucokinase in complex with a synthetic activator | | Descriptor: | 1-({5-[4-(methylsulfonyl)phenoxy]-2-pyridin-2-yl-1H-benzimidazol-6-yl}methyl)pyrrolidine-2,5-dione, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K, Takahashi, K. | | Deposit date: | 2009-04-14 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The design and optimization of a series of 2-(pyridin-2-yl)-1H-benzimidazole compounds as allosteric glucokinase activators.
Bioorg.Med.Chem., 17, 2009
|
|
3A0I
 
 | | Human glucokinase in complex with a synthetic activator | | Descriptor: | 3-[(4-fluorophenyl)sulfanyl]-N-(4-methyl-1,3-thiazol-2-yl)-6-[(4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl]pyridine-2-carboxamide, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K, Mitsuya, M. | | Deposit date: | 2009-03-19 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel 3,6-disubstituted 2-pyridinecarboxamide derivatives as GK activators
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5K7K
 
 | |
3FR0
 
 | | Human glucokinase in complex with 2-amino benzamide activator | | Descriptor: | 2-amino-N-(4-methyl-1,3-thiazol-2-yl)-5-[(4-methyl-4H-1,2,4-triazol-3-yl)sulfanyl]benzamide, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K. | | Deposit date: | 2009-01-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of novel and potent 2-amino benzamide derivatives as allosteric glucokinase activators
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3WXA
 
 | | X-ray crystal structural analysis of the complex between ALG-2 and Sec31A peptide | | Descriptor: | Programmed cell death protein 6, Protein transport protein Sec31A, ZINC ION | | Authors: | Takahashi, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2
Int J Mol Sci, 16, 2015
|
|
