1B90
 
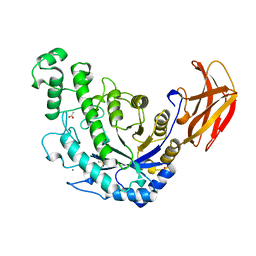 | | BACILLUS CEREUS BETA-AMYLASE APO FORM | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1B9Z
 
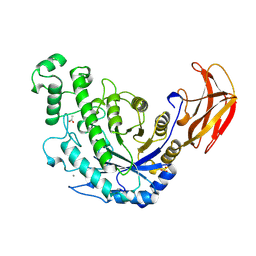 | | BACILLUS CEREUS BETA-AMYLASE COMPLEXED WITH MALTOSE | | Descriptor: | ACETATE ION, CALCIUM ION, PROTEIN (BETA-AMYLASE), ... | | Authors: | Mikami, B, Adachi, M, Kage, T, Sarikaya, E, Nanmori, T, Shinke, R, Utsumi, S. | | Deposit date: | 1999-03-06 | | Release date: | 1999-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of raw starch-digesting Bacillus cereus beta-amylase complexed with maltose.
Biochemistry, 38, 1999
|
|
1NNX
 
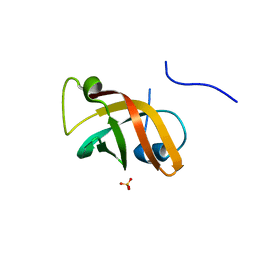 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
1RXX
 
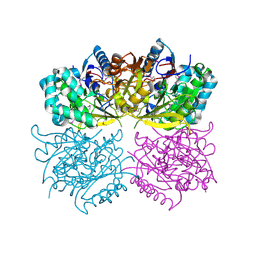 | | Structure of arginine deiminase | | Descriptor: | Arginine deiminase | | Authors: | Galkin, A, Kulakova, L, Sarikaya, E, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target.
J.Biol.Chem., 279, 2004
|
|
1IZM
 
 | | Structure of ygfB from Haemophilus influenzae (HI0817), a Conserved Hypothetical Protein | | Descriptor: | HYPOTHETICAL PROTEIN HI0817 | | Authors: | Galkin, A, Sarikaya, E, Lehmann, C, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-09 | | Release date: | 2003-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | X-ray structure of HI0817 from Haemophilus influenzae: protein of unknown function with a novel fold.
Proteins, 57, 2004
|
|
1MW5
 
 | | Structure of HI1480 from Haemophilus influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI1480 | | Authors: | Lim, K, Sarikaya, E, Howard, A, Galkin, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-09-27 | | Release date: | 2003-11-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1NU0
 
 | | Structure of the double mutant (L6M; F134M, SeMet form) of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF, SULFATE ION | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-30 | | Release date: | 2004-03-02 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
1NMN
 
 | | Structure of yqgF from Escherichia coli, a hypothetical protein | | Descriptor: | Hypothetical protein yqgF | | Authors: | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
2FH8
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with isomaltose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-6)-beta-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FHC
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltotriose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FH6
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with glucose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FHB
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FGZ
 
 | | Crystal Structure Analysis of apo pullulanase from Klebsiella pneumoniae | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FHF
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltotetraose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
1NRK
 
 | |
