1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
1Q0B
 
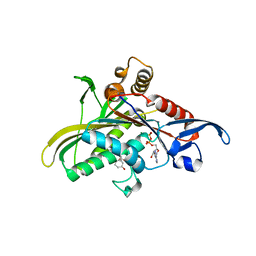 | | Crystal structure of the motor protein KSP in complex with ADP and monastrol | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHYL 4-(3-HYDROXYPHENYL)-6-METHYL-2-THIOXO-1,2,3,4-TETRAHYDROPYRIMIDINE-5-CARBOXYLATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y, Sardana, V, Xu, B, Halczenko, W, Homnick, C, Buser, C.A, Hartman, G.D, Huber, H.E, Kuo, L.C. | | Deposit date: | 2003-07-15 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of a mitotic motor protein: where, how, and conformational consequences
J.Mol.Biol., 335, 2004
|
|
1JXP
 
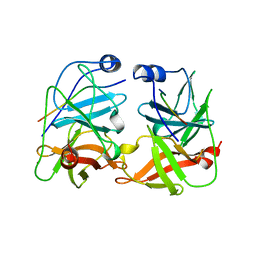 | | BK STRAIN HEPATITIS C VIRUS (HCV) NS3-NS4A | | Descriptor: | NS3 SERINE PROTEASE, NS4A, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-08-21 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
2A4R
 
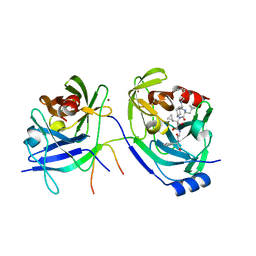 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4Q
 
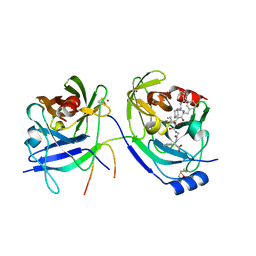 | | HCV NS3 protease with NS4a peptide and a covalently bound macrocyclic ketoamide compound. | | Descriptor: | (2R)-({N-[(3S)-3-({[(3S,6S)-6-CYCLOHEXYL-5,8-DIOXO-4,7-DIAZABICYCLO[14.3.1]ICOSA-1(20),16,18-TRIEN-3-YL]CARBONYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, NS3 protease/helicase', ... | | Authors: | Chen, K.X, Njoroge, F.G, Prongay, A, Pichardo, J, Madison, V, Girijavallabhan, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4G
 
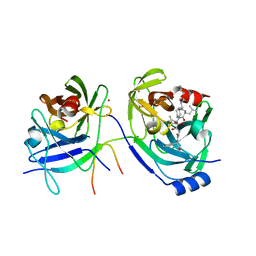 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1NS3
 
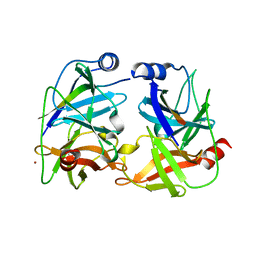 | | STRUCTURE OF HCV PROTEASE (BK STRAIN) | | Descriptor: | NS3 PROTEASE, NS4A PEPTIDE, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-04-05 | | Release date: | 1998-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
1BT7
 
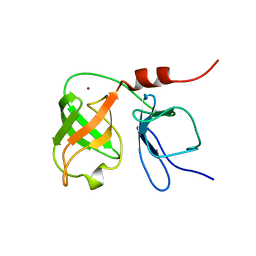 | | THE SOLUTION NMR STRUCTURE OF THE N-TERMINAL PROTEASE DOMAIN OF THE HEPATITIS C VIRUS (HCV) NS3-PROTEIN, FROM BK STRAIN, 20 STRUCTURES | | Descriptor: | NS3 SERINE PROTEASE, ZINC ION | | Authors: | Barbato, G, Cicero, D.O, Nardi, M.C, Steinkuhler, C, Cortese, R, De Francesco, R, Bazzo, R. | | Deposit date: | 1998-09-01 | | Release date: | 1999-06-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal proteinase domain of the hepatitis C virus (HCV) NS3 protein provides new insights into its activation and catalytic mechanism.
J.Mol.Biol., 289, 1999
|
|
3LP3
 
 | | p15 HIV RNaseH domain with inhibitor MK3 | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, p15 | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3CUK
 
 | |
3F9N
 
 | | Crystal structure of chk1 kinase in complex with inhibitor 38 | | Descriptor: | 3-(3-chlorophenyl)-2-({(1S)-1-[(6S)-2,8-diazaspiro[5.5]undec-2-ylcarbonyl]pentyl}sulfanyl)quinazolin-4(3H)-one, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y, Munshi, S, Ikuta, M. | | Deposit date: | 2008-11-14 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of thioquinazolinones, allosteric Chk1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1TCP
 
 | |
1TQF
 
 | | Crystal structure of human Beta secretase complexed with inhibitor | | Descriptor: | 3-{2-[(5-AMINOPENTYL)AMINO]-2-OXOETHOXY}-5-({[1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)PHENYL PHENYLMETHANESULFONATE, Beta-secretase 1 | | Authors: | Munshi, S, Chen, Z, Kuo, L. | | Deposit date: | 2004-06-17 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a small molecule nonpeptide active site beta-secretase inhibitor that displays a nontraditional binding mode for aspartyl proteases.
J.Med.Chem., 47, 2004
|
|
3LP2
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 3-[4-(diethylamino)phenoxy]-6-(ethoxycarbonyl)-5,8-dihydroxy-7-oxo-7,8-dihydro-1,8-naphthyridin-1-ium, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP1
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 3-cyclopentyl-1,4-dihydroxy-1,8-naphthyridin-2(1H)-one, MANGANESE (II) ION, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
3LP0
 
 | | HIV-1 reverse transcriptase with inhibitor | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Yan, Y, Munshi, S.K, Prasad, G.S, Su, H.P. | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors.
J.Virol., 84, 2010
|
|
2HXL
 
 | | crystal structure of Chek1 in complex with inhibitor 1 | | Descriptor: | 3-(5-{[4-(AMINOMETHYL)PIPERIDIN-1-YL]METHYL}-1H-INDOL-2-YL)-1H-INDAZOLE-6-CARBONITRILE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 6-substituted indolylquinolinones as potent Chek1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HY0
 
 | | crystal structure of chek1 in complex with inhibitor 22 | | Descriptor: | 3-[5-(PIPERIDIN-1-YLMETHYL)-1H-INDOL-2-YL]-6-(1H-PYRAZOL-4-YL)QUINOLIN-2(1H)-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y. | | Deposit date: | 2006-08-04 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of 6-substituted indolylquinolinones as potent Chek1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2HXQ
 
 | | crystal structure of Chek1 in complex with inhibitor 2 | | Descriptor: | 3-(5-{[4-(AMINOMETHYL)PIPERIDIN-1-YL]METHYL}-1H-INDOL-2-YL)QUINOLIN-2(1H)-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Yan, Y. | | Deposit date: | 2006-08-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of 6-substituted indolylquinolinones as potent Chek1 kinase inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
