4W8O
 
 | | Structure of the luciferase-like enzyme from the nonluminescent Zophobas morio mealworm | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, luciferase-like enzymeAMP-CoA-ligase | | Authors: | Santos, C.R, Prado, R.A, Viviani, V, Murakami, M.T. | | Deposit date: | 2014-08-25 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the luciferase-like enzyme from the nonluminescent Zophobas morio mealworm
To Be Published
|
|
3H79
 
 | | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70 | | Descriptor: | THIOCYANATE ION, Thioredoxin-like protein | | Authors: | Santos, C.R, Fessel, M.R, Vieira, L.C, Krieger, M.A, Goldenberg, S, Guimaraes, B.G, Zanchin, N.I.T, Barbosa, J.A.R.G. | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Trypanosoma cruzi thioredoxin-like hypothetical protein Q4DV70
TO BE PUBLISHED
|
|
4KCA
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from a Bovine Ruminal Metagenomic Library | | Descriptor: | Endo-1,5-alpha-L-arabinanase, GLYCEROL, IODIDE ION, ... | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4KCB
 
 | | Crystal Structure of Exo-1,5-alpha-L-arabinanase from Bovine Ruminal Metagenomic Library | | Descriptor: | Arabinan endo-1,5-alpha-L-arabinosidase, PHOSPHATE ION | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2014-04-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
6UB3
 
 | | Crystal structure of a GH128 (subgroup IV) endo-beta-1,3-glucanase from Lentinula edodes (LeGH128_IV) with laminaribiose at the surface-binding site | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Santos, C.R, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
4PN2
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylotriose | | Descriptor: | CALCIUM ION, Xylanase, beta-D-xylopyranose | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMY
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylose | | Descriptor: | CALCIUM ION, GLYCEROL, Xylanase, ... | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMX
 
 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri in the native form | | Descriptor: | CALCIUM ION, Xylanase | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.304 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMV
 
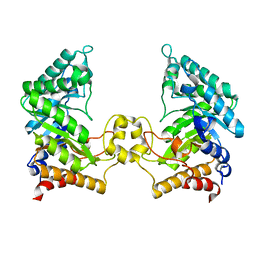 | | Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P43212) | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMU
 
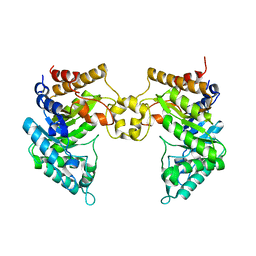 | | Crystal structure of a novel reducing-end xylose-releasing exo-oligoxylanase (XynA) belonging to GH10 family (space group P1211) | | Descriptor: | Endo-1,4-beta-xylanase A | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.857 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
4PMZ
 
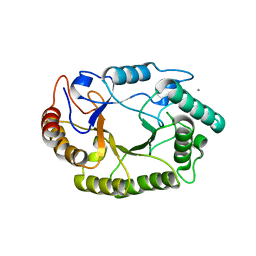 | | Crystal structure of GH10 endo-b-1,4-xylanase (XynB) from Xanthomonas axonopodis pv citri complexed with xylobiose | | Descriptor: | CALCIUM ION, Xylanase, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Santos, C.R, Martins, V.P.M, Zanphorlin, L.M, Ruller, R, Murakami, M.T. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Molecular mechanisms associated with xylan degradation by xanthomonas plant pathogens.
J.Biol.Chem., 289, 2014
|
|
7UFR
 
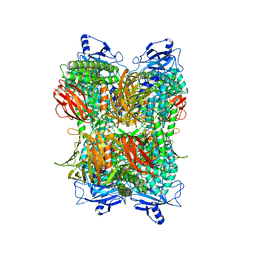 | | Cryo-EM Structure of Bl_Man38A at 2.7 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structure of Bl_Man38A at 2.7 A
Nat.Chem.Biol., 2022
|
|
7UFU
 
 | | Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A | | Descriptor: | Alpha-mannosidase, ZINC ION, alpha-D-mannopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A
Nat.Chem.Biol., 2022
|
|
7UFT
 
 | | Cryo-EM Structure of Bl_Man38C at 2.9 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structure of Bl_Man38C at 2.9 A
Nat.Chem.Biol., 2022
|
|
7UFS
 
 | | Cryo-EM Structure of Bl_Man38B at 3.4 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of Bl_Man38B at 3.4 A
Nat.Chem.Biol., 2022
|
|
4EKJ
 
 | | Crystal structure of a monomeric beta-xylosidase from Caulobacter crescentus CB15 | | Descriptor: | Beta-xylosidase, SULFATE ION | | Authors: | Santos, C.R, Polo, C.C, Correa, J.M, Simao, R.C.G, Seixas, F.A.V, Murakami, M.T. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The accessory domain changes the accessibility and molecular topography of the catalytic interface in monomeric GH39 beta-xylosidases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3N98
 
 | | Crystal structure of TK1436, a GH57 branching enzyme from hyperthermophilic archaeon Thermococcus kodakaraensis, in complex with glucose and additives | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santos, C.R, Tonoli, C.C.C, Trindade, D.M, Betzel, C, Takata, H, Kuriki, T, Kanai, T, Imanaka, T, Arni, R.K, Murakami, M.T. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for branching-enzyme activity of glycoside hydrolase family 57: Structure and stability studies of a novel branching enzyme from the hyperthermophilic archaeon Thermococcus Kodakaraensis KOD1.
Proteins, 79, 2011
|
|
3N92
 
 | | Crystal structure of TK1436, a GH57 branching enzyme from hyperthermophilic archaeon Thermococcus kodakaraensis, in complex with glucose | | Descriptor: | alpha-amylase, GH57 family, beta-D-glucopyranose | | Authors: | Santos, C.R, Tonoli, C.C.C, Trindade, D.M, Betzel, C, Takata, H, Kuriki, T, Kanai, T, Imanaka, T, Arni, R.K, Murakami, M.T. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural basis for branching-enzyme activity of glycoside hydrolase family 57: Structure and stability studies of a novel branching enzyme from the hyperthermophilic archaeon Thermococcus Kodakaraensis KOD1.
Proteins, 79, 2011
|
|
3N8T
 
 | | Native structure of TK1436, a GH57 branching enzyme from hyperthermophilic archaeon Thermococcus kodakaraensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, alpha-amylase, ... | | Authors: | Santos, C.R, Tonoli, C.C.C, Trindade, D.M, Betzel, C, Takata, H, Kuriki, T, Kanai, T, Imanaka, T, Arni, R.K, Murakami, M.T. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for branching-enzyme activity of glycoside hydrolase family 57: Structure and stability studies of a novel branching enzyme from the hyperthermophilic archaeon Thermococcus Kodakaraensis KOD1.
Proteins, 79, 2011
|
|
3NIY
 
 | | Crystal structure of native xylanase 10B from Thermotoga petrophila RKU-1 | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Santos, C.R, Meza, A.N, Trindade, D.M, Ruller, R, Squina, F.M, Prade, R.A, Murakami, M.T. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Thermal-induced conformational changes in the product release area drive the enzymatic activity of xylanases 10B: Crystal structure, conformational stability and functional characterization of the xylanase 10B from Thermotoga petrophila RKU-1.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
3NJ3
 
 | | Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Santos, C.R, Meza, A.N, Trindade, D.M, Ruller, R, Squina, F.M, Prade, R.A, Murakami, M.T. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Thermal-induced conformational changes in the product release area drive the enzymatic activity of xylanases 10B: Crystal structure, conformational stability and functional characterization of the xylanase 10B from Thermotoga petrophila RKU-1.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
3PZM
 
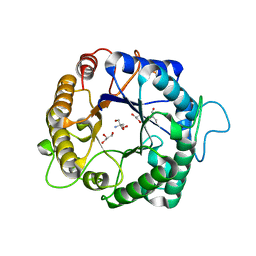 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5 | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZ9
 
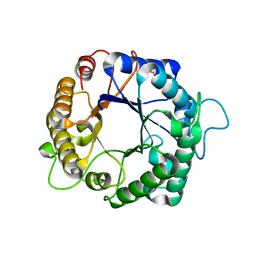 | | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 | | Descriptor: | Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5 | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZO
 
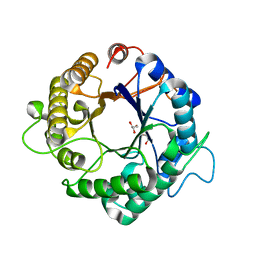 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules | | Descriptor: | GLYCEROL, Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
3PZU
 
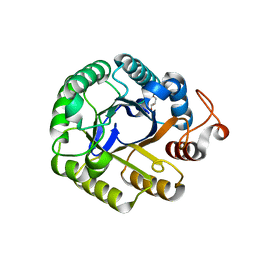 | | P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 | | Descriptor: | Endoglucanase, GLYCEROL | | Authors: | Santos, C.R, Paiva, J.H, Akao, P.K, Meza, A.N, Silva, J.C, Squina, F.M, Ward, R.J, Ruller, R, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168.
Biochem.J., 441, 2012
|
|
