2KG4
 
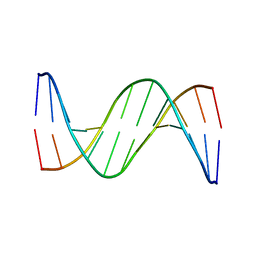
 | | Three-dimensional structure of human Gadd45alpha in solution by NMR | | Descriptor: | Growth arrest and DNA-damage-inducible protein GADD45 alpha | | Authors: | Sanchez, R, Pantoja-Uceda, D, Prieto, J, Diercks, T, Campos-Olivas, R, Blanco, F.J. | | Deposit date: | 2009-03-04 | | Release date: | 2009-03-31 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human growth arrest and DNA damage 45alpha (Gadd45alpha) and its interactions with proliferating cell nuclear antigen (PCNA) and Aurora A kinase
J.Biol.Chem., 285, 2010
|
|
1ZS5
 
 | | Structure-based evaluation of selective and non-selective small molecules that block HIV-1 TAT and PCAF association | | Descriptor: | (3E)-4-(1-METHYL-1H-INDOL-3-YL)BUT-3-EN-2-ONE, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Godbole, S, Muller, M, Yan, S, Sanchez, R, Zhou, M. | | Deposit date: | 2005-05-23 | | Release date: | 2006-05-23 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure-based evaluation of selective nad non-selective small molecules that block hiv-1 tat and pcaf association
TO BE PUBLISHED
|
|
6AU2
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4H,6H-[1,2,4]triazolo[4,3-a][4,1]benzoxazepine, BETA-MERCAPTOETHANOL, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-10-11 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification and characterization of the first fragment hits for SETDB1 Tudor domain.
Bioorg.Med.Chem., 27, 2019
|
|
6AU3
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, N-{[2-(3,5-dimethyl-4H-1,2,4-triazol-4-yl)phenyl]methyl}acetamide, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, IQBAL, A, DONG, A, DOBROVETSKY, E, CORLESS, V.B, LIEW, S.K, TEMPEL, W, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D, HOLOWNIA, A, VEDADI, M, BROWN, P.J, SANTHAKUMAR, V, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-08-30 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragments
to be published
|
|
6BPI
 
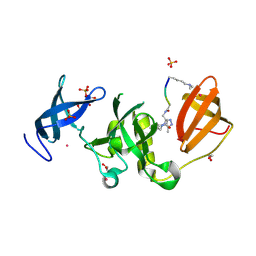 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, MLY-SER-THR-E2G, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, DONG, A, DOBROVETSKY, E, IQBAL, A, CORLESS, V, TEMPEL, W, LIEW, S.K, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D.B, SCHAPIRA, M, VEDADI, M, BROWN, P.J, Santhakumar, V, FRYE, S, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates
to be published
|
|
6RZU
 
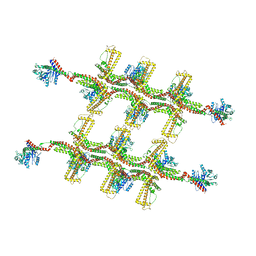 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZT
 
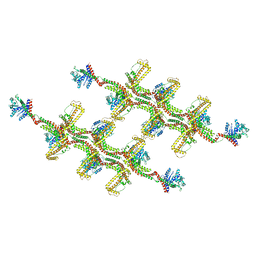 | | Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (14.7 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZV
 
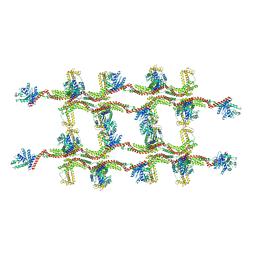 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (20.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
6RZW
 
 | | Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state | | Descriptor: | Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Sanchez, R, Kudryashev, M, Kuelbrandt, W, Daumke, O. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-24 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (18.799999 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
2WBB
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-BROMO-1,3-THIAZOL-2(3H)-YLIDENE]CARBAMOYL}-4-METHYLBENZENESULFONAMIDE | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
2WBD
 
 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE-1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH AN AMP SITE INHIBITOR | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-3-ethylbenzenesulfonamide | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T, Kitas, E, Mohr, P, Kuhn, B, Wessel, H.P, Hebeisen, P, Haap, W, Huber, W, Alvarez Sanchez, R, Paehler, A, Bernadeau, A, Gubler, M, Schott, B, Tozzo, E. | | Deposit date: | 2009-02-26 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Sulfonylureido Thiazoles as Fructose-1,6-Bisphosphatase Inhibitors for the Treatment of Type-2 Diabetes.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6QL4
 
 | | Crystal structure of nucleotide-free Mgm1 | | Descriptor: | 1,2-ETHANEDIOL, Putative mitochondrial dynamin protein | | Authors: | Faelber, K, Dietrich, L, Noel, J.K, Wollweber, F, Pfitzner, A.-K, Muehleip, A, Sanchez, R, Kudryashev, M, Chiaruttin, N, Lilie, H, Schlegel, J, Rosenbaum, E, Hessenberger, M, Matthaeus, C, Noe, F, Roux, A, vanderLaan, M, Kuehlbrandt, W, Daumke, O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1.
Nature, 571, 2019
|
|
2Y5L
 
 | | orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-bromo-1,3-thiazol-2(3H)-ylidene]carbamoyl}-3-chlorobenzenesulfonamide | | Authors: | ruf, a, hebeisen, p, haap, w, kuhn, b, mohr, p, wessel, h.p, zutter, u, kirchner, s, benz, j, joseph, c, alvarez-sanchez, r, gubler, m, schott, b, benardeau, a, tozzo, e, kitas, e. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y5K
 
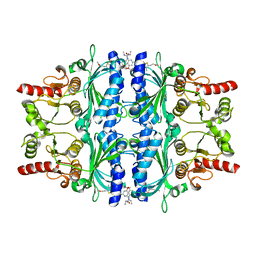 | | Orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | 1-[5-(2-METHOXYETHYL)-4-METHYL-THIOPHEN-2-YL]SULFONYL-3-[4-METHOXY-6-(METHYLCARBAMOYLAMINO)PYRIDIN-2-YL]UREA, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Hebeisen, P, Haap, W, Kuhn, B, Mohr, P, Wessel, H.P, Zutter, U, Kirchner, S, Benz, J, Joseph, C, Alvarez-Sanchez, R, Gubler, M, Schott, B, Benardeau, A, Tozzo, E, Kitas, E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4J2I
 
 | | Multiple crystal structures of an all-AT DNA dodecamer stabilized by weak interactions | | Descriptor: | 5'-D(*AP*AP*TP*AP*AP*AP*TP*TP*TP*AP*TP*T)-3' | | Authors: | Acosta-Reyes, F.J, Subirana, J.A, Pous, J, Sanchez, R, Condom, N, Baldini, R, Malinina, L, Campos, J.L. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Polymorphic crystal structures of an all-AT DNA dodecamer.
Biopolymers, 103, 2015
|
|
4AIX
 
 | | Crystallographic structure of an amyloidogenic variant, 3rC34Y, of the germinal line lambda 3 | | Descriptor: | IG LAMBDA CHAIN V-IV REGION BAU | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
4AJ0
 
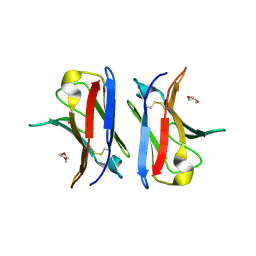 | | Crystallographic structure of an amyloidogenic variant, 3rCW, of the germinal line lambda 3 | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, ACETATE ION, GERMINAL LINE LAMBDA 3 3RCW VARIANT | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
4AIZ
 
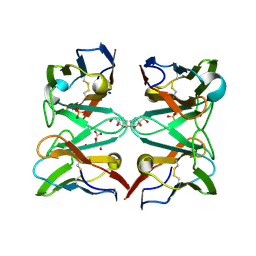 | | Crystallographic structure of 3mJL2 from the germinal line lambda 3 | | Descriptor: | 1,5:6,10-dianhydro-3,4,7,8-tetradeoxy-2,9-bis-C-(hydroxymethyl)-L-manno-decitol, CITRIC ACID, SULFATE ION, ... | | Authors: | Villalba, M.I, Luna, O.D, Rudino-Pinera, E, Sanchez, R, Sanchez-Lopez, R, Rojas-Trejo, S, Olamendi-Portugal, T, Fernandez-Velasco, D.A, Becerril, B. | | Deposit date: | 2012-02-15 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-Directed Mutagenesis Reveals Regions Implicated in the Stability and Fiber Formation of Human Lambda3R Light Chains.
J.Biol.Chem., 290, 2015
|
|
3I0R
 
 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 3 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6-methyl-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3I0S
 
 | | crystal structure of HIV reverse transcriptase in complex with inhibitor 7 | | Descriptor: | Reverse transcriptase/ribonuclease H, S-{2-[(2-chloro-4-sulfamoylphenyl)amino]-2-oxoethyl} 6,8-dichloro-3,4-dihydroquinoline-1(2H)-carbothioate, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substituted tetrahydroquinolines as potent allosteric inhibitors of reverse transcriptase and its key mutants.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8C8J
 
 | | Long Interspersed Nuclear Element 1 (LINE-1) reverse transcriptase ternary complex with hybrid duplex and dTTP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, CHLORIDE ION, ... | | Authors: | Nichols, C.E, Walpole, T.B, Baldwin, E. | | Deposit date: | 2023-01-20 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures, functions and adaptations of the human LINE-1 ORF2 protein.
Nature, 626, 2024
|
|
3C6T
 
 | | Crystal Structure of HIV Reverse Transcriptase in complex with inhibitor 14 | | Descriptor: | 2-[3-chloro-5-(3-chloro-5-cyanophenoxy)phenoxy]-N-(2-chloro-4-sulfamoylphenyl)acetamide, Reverse transcriptase | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3C6U
 
 | | Crystal Structure of HIV Reverse Transcriptase in complex with inhibitor 22 | | Descriptor: | 3-chloro-5-[2-chloro-5-(1H-indazol-3-ylmethoxy)phenoxy]benzonitrile, Reverse transcriptase | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-02-05 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The design and synthesis of diaryl ether second generation HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTIs) with enhanced potency versus key clinical mutations.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DRP
 
 | | HIV reverse transcriptase in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y, Prasad, S. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3DRR
 
 | | HIV reverse transcriptase Y181C mutant in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
