1JU8
 
 | | Solution structure of Leginsulin, a plant hormon | | Descriptor: | Leginsulin | | Authors: | Yamazaki, T, Takaoka, M, Katoh, E, Hanada, K, Sakita, M, Sakata, K, Nishiuchi, Y, Hirano, H. | | Deposit date: | 2001-08-23 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A possible physiological function and the tertiary structure of a 4-kDa peptide in legumes
EUR.J.BIOCHEM., 270, 2003
|
|
1VBH
 
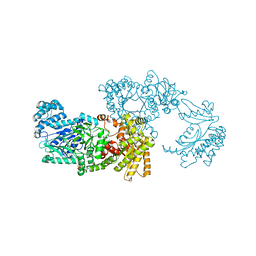 | | Pyruvate Phosphate Dikinase with bound Mg-PEP from Maize | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, SULFATE ION, ... | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1VBG
 
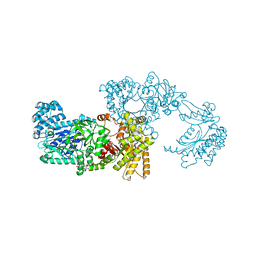 | | Pyruvate Phosphate Dikinase from Maize | | Descriptor: | MAGNESIUM ION, SULFATE ION, pyruvate,orthophosphate dikinase | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
2D1Q
 
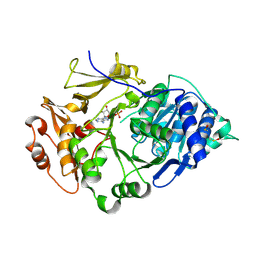 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with MgATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1R
 
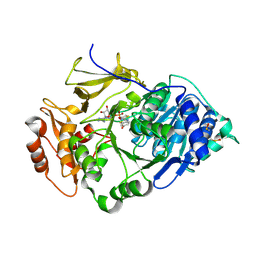 | | Crystal structure of the thermostable Japanese firefly Luciferase complexed with OXYLUCIFERIN and AMP | | Descriptor: | 2-(6-HYDROXY-1,3-BENZOTHIAZOL-2-YL)-1,3-THIAZOL-4(5H)-ONE, ADENOSINE MONOPHOSPHATE, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1S
 
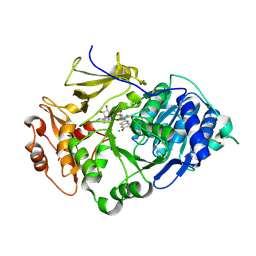 | | Crystal structure of the thermostable Japanese Firefly Luciferase complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
2D1T
 
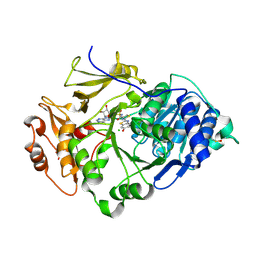 | | Crystal structure of the thermostable Japanese Firefly Luciferase red-color emission S286N mutant complexed with High-energy intermediate analogue | | Descriptor: | 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, Luciferin 4-monooxygenase | | Authors: | Nakatsu, T, Ichiyama, S, Hiratake, J, Saldanha, A, Kobashi, N, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-31 | | Release date: | 2006-03-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for the spectral difference in luciferase bioluminescence.
Nature, 440, 2006
|
|
5WR7
 
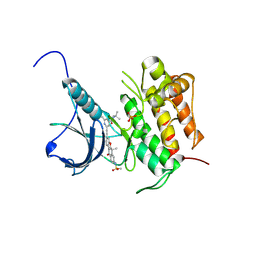 | | Crystal structure of Trk-A complexed with a selective inhibitor CH7057288 | | Descriptor: | High affinity nerve growth factor receptor, N-tert-butyl-2-[2-[6,6-dimethyl-8-(methylsulfonylamino)-11-oxidanylidene-naphtho[2,3-b][1]benzofuran-3-yl]ethynyl]-6-methyl-pyridine-4-carboxamide | | Authors: | Tanaka, H, Blaesse, M, Augustin, M, Goesser, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-12-06 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Selective TRK Inhibitor CH7057288 against TRK Fusion-Driven Cancer.
Mol. Cancer Ther., 17, 2018
|
|
5B7V
 
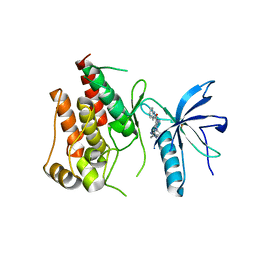 | | Human FGFR1 kinase in complex with CH5183284 | | Descriptor: | Fibroblast growth factor receptor 1, SULFATE ION, [5-amino-1-(2-methyl-1H-benzimidazol-6-yl)-1H-pyrazol-4-yl](1H-indol-2-yl)methanone | | Authors: | Fukami, T.A, Lukacs, C.M, Janson, C. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The fibroblast growth factor receptor genetic status as a potential predictor of the sensitivity to CH5183284/Debio 1347, a novel selective FGFR inhibitor
Mol.Cancer Ther., 13, 2014
|
|
1IYK
 
 | | Crystal structure of candida albicans N-myristoyltransferase with myristoyl-COA and peptidic inhibitor | | Descriptor: | MYRISTOYL-COA:PROTEIN N-MYRISTOYLTRANSFERASE, TETRADECANOYL-COA, [CYCLOHEXYLETHYL]-[[[[4-[2-METHYL-1-IMIDAZOLYL-BUTYL]PHENYL]ACETYL]-SERYL]-LYSINYL]-AMINE | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
1IYL
 
 | | Crystal Structure of Candida albicans N-myristoyltransferase with Non-peptidic Inhibitor | | Descriptor: | (1-METHYL-1H-IMIDAZOL-2-YL)-(3-METHYL-4-{3-[(PYRIDIN-3-YLMETHYL)-AMINO]-PROPOXY}-BENZOFURAN-2-YL)-METHANONE, Myristoyl-CoA:Protein N-Myristoyltransferase | | Authors: | Sogabe, S, Fukami, T.A, Morikami, K, Shiratori, Y, Aoki, Y, D'Arcy, A, Winkler, F.K, Banner, D.W, Ohtsuka, T. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structures of Candida albicans N-Myristoyltransferase with Two Distinct Inhibitors
CHEM.BIOL., 9, 2002
|
|
7E18
 
 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor YH-53 | | Descriptor: | 1,2-ETHANEDIOL, N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Replicase polyprotein 1ab | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
7E19
 
 | | Crystal structure of SAR-CoV-2 3CL protease complex with inhibitor SH-5 | | Descriptor: | (phenylmethyl) N-[(2S)-1-[[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]carbamate, 3C-like proteinase | | Authors: | Senda, M, Konno, S, Hayashi, Y, Senda, T. | | Deposit date: | 2021-02-01 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents.
J.Med.Chem., 65, 2022
|
|
3WQ4
 
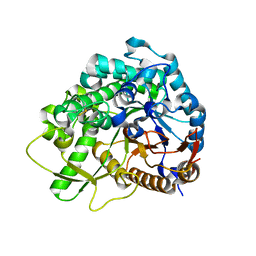 | | Crystal structure of beta-primeverosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-primeverosidase | | Authors: | Saino, H. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of beta-primeverosidase in complex with disaccharide amidine inhibitors.
J.Biol.Chem., 289, 2014
|
|
3WQ5
 
 | |
3WQ6
 
 | |
2ZXQ
 
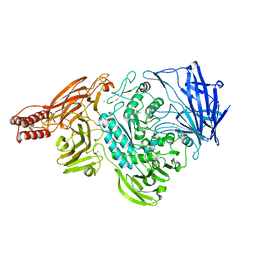 | | Crystal structure of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Endo-alpha-N-acetylgalactosaminidase, MANGANESE (II) ION | | Authors: | Suzuki, R, Katayama, T, Ashida, H, Yamamoto, K, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-06-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational analyses of substrate recognition of endo-{alpha}-N-acetylgalactosaminidase from Bifidobacterium longum.
J.Biochem., 2009
|
|
