5X5H
 
 | | Crystal structure of metB from Corynebacterium glutamicum | | Descriptor: | Cystathionine beta-lyases/cystathionine gamma-synthases, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Insights into Substrate Specificity of Cystathionine gamma-Synthase from Corynebacterium glutamicum
J. Agric. Food Chem., 65, 2017
|
|
7CBE
 
 | |
6JTT
 
 | | MHETase in complex with BHET | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, Mono(2-hydroxyethyl) terephthalate hydrolase, ... | | Authors: | Sagong, H.-Y, Seo, H, Kim, K.-J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Decomposition of PET film by MHETase using Exo-PETase function
Acs Catalysis, 10, 2020
|
|
6JTU
 
 | | Crystal structure of MHETase from Ideonella sakaiensis | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Sagong, H.-Y, Seo, H, Kim, K.-J. | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Decomposition of PET film by MHETase using Exo-PETase function
Acs Catalysis, 10, 2020
|
|
5GJO
 
 | |
5GJN
 
 | |
5GJM
 
 | |
5GJP
 
 | |
5M47
 
 | |
7D7O
 
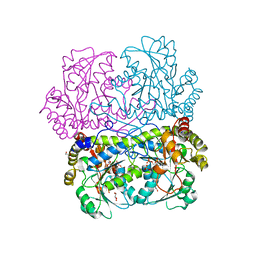 | | Crystal structure of cystathionine gamma-lyase from Bacillus cereus ATCC 14579 | | Descriptor: | Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sagong, H.-Y, Kim, B, Kim, K.-J. | | Deposit date: | 2020-10-05 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural and Functional Characterization of Cystathionine gamma-lyase from Bacillus cereus ATCC 14579.
J.Agric.Food Chem., 68, 2020
|
|
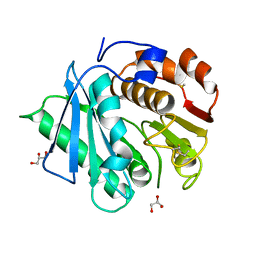
7DZV
 
 | |
7DZT
 
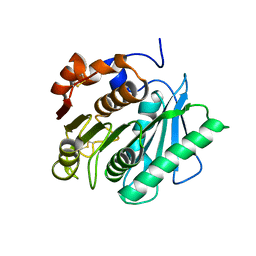 | | Cyrstal structure of PETase from Rhizobacter gummiphilus | | Descriptor: | DLH domain-containing protein | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Implications for the PET decomposition mechanism through similarity and dissimilarity between PETases from Rhizobacter gummiphilus and Ideonella sakaiensis.
J Hazard Mater, 416, 2021
|
|
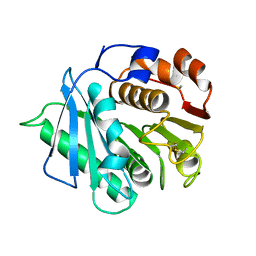
7DZU
 
 | |
6IOI
 
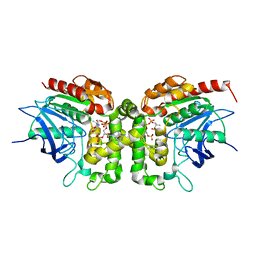 | |
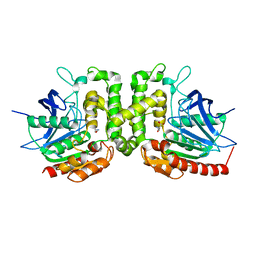
6IOH
 
 | |
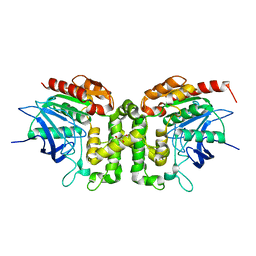
6IOG
 
 | |
5E3Q
 
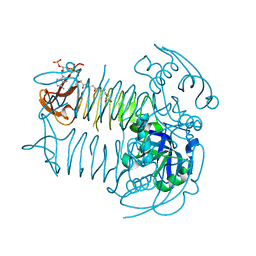 | | Crystal structure of DapD in complex with succinyl-CoA from Corynebacterium glutamicum | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase, SUCCINYL-COENZYME A | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2015-10-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure and Biochemical Characterization of Tetrahydrodipicolinate N-Succinyltransferase from Corynebacterium glutamicum.
J.Agric.Food Chem., 63, 2015
|
|
5E3R
 
 | | Crystal structure of DapD in complex with 2-aminopimelate from Corynebacterium glutamicum | | Descriptor: | (2S)-2-aminoheptanedioic acid, 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2015-10-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure and Biochemical Characterization of Tetrahydrodipicolinate N-Succinyltransferase from Corynebacterium glutamicum.
J.Agric.Food Chem., 63, 2015
|
|
5E3P
 
 | | Crystal structure of DapD from Corynebacterium glutamicum | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-succinyltransferase, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2015-10-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure and Biochemical Characterization of Tetrahydrodipicolinate N-Succinyltransferase from Corynebacterium glutamicum.
J.Agric.Food Chem., 63, 2015
|
|
5EER
 
 | | Crystal structure of DapB from Corynebacterium glutamicum | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, SULFATE ION | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2015-10-23 | | Release date: | 2015-11-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insight into Dihydrodipicolinate Reductase from Corybebacterium glutamicum for Lysine Biosynthesis.
J. Microbiol. Biotechnol., 26, 2016
|
|
5EES
 
 | |
5H2Y
 
 | |
5H2G
 
 | |
5YNS
 
 | |
5XJH
 
 | |
