6R8Q
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A BENZOTRIAZOLE FRAGMENT | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R.R, Vecchia, L, Jones, E.Y. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
6YUW
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 454 | | Descriptor: | 1-(cyclopropylmethyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV2
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 598 | | Descriptor: | (3~{R})-1-phenylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV0
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 587 | | Descriptor: | (3~{R})-1-(2-chlorophenyl)pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6Z43
 
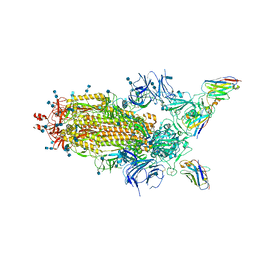 | | Cryo-EM Structure of SARS-CoV-2 Spike : H11-D4 Nanobody Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody, ... | | Authors: | Ruza, R.R, Duyvesteyn, H.M.E, Shah, P, Carrique, L, Ren, J, Malinauskas, T, Zhou, D, Stuart, D.I, Naismith, J.H. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for a potent neutralising single-domain antibody that blocks SARS-CoV-2 binding to its receptor ACE2
To Be Published
|
|
6R8R
 
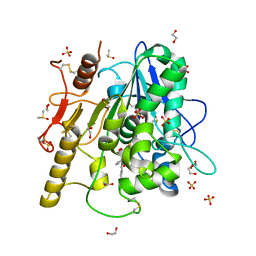 | | Structure of the Wnt deacylase Notum in complex with isoquinoline 45 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Zhao, Y, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
5L4H
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with 5-(2-BROMO-ETHYL)-5-ETHYL-PYRIMIDINE-2,4,6-TRIONE (brominated barbiturate) | | Descriptor: | 5-(2-bromoethyl)-5-ethyl-1,3-diazinane-2,4,6-trione, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2017-02-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
5L4E
 
 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with thiopental | | Descriptor: | 5-ethyl-5-[(2R)-pentan-2-yl]-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, CHLORIDE ION, DODECANE, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2017-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
5MUO
 
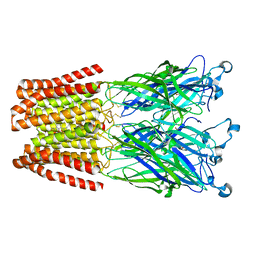 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Fourati, Z, Ruza, R.R, Delarue, M. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
6YOR
 
 | | Structure of the SARS-CoV-2 spike S1 protein in complex with CR3022 Fab | | Descriptor: | IgG H chain, IgG L chain, Spike glycoprotein | | Authors: | Huo, J, Zhao, Y, Ren, J, Zhou, D, Duyvesteyn, H.M.E, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-04-15 | | Release date: | 2020-04-29 | | Last modified: | 2022-04-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6YXI
 
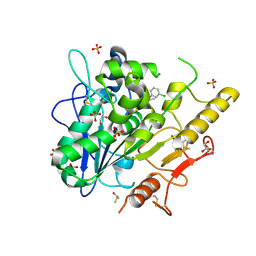 | | Structure of Notum in complex with a 1-(3-Chlorophenyl)-2,5-dimethyl-1H-pyrrole-3-carboxylic acid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-chlorophenyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchia, L, Jones, E.Y, Ruza, R.R, Hillier, J, Zhao, Y. | | Deposit date: | 2020-05-01 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV4
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 686 | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YUY
 
 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 471 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-methyl-5-(trifluoromethyl)-1~{H}-pyrrole-3-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J, Ruza, R.R, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6Z97
 
 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-06-03 | | Release date: | 2020-07-01 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
7PKV
 
 | | Notum_Inhibitor ARUK3000223 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R, Zhao, Y, Fish, P, Jones, E.Y. | | Deposit date: | 2021-08-26 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Design of a Potent, Selective, and Brain-Penetrant Inhibitor of Wnt-Deactivating Enzyme Notum by Optimization of a Crystallographic Fragment Hit.
J.Med.Chem., 65, 2022
|
|
6EMX
 
 | |
5L47
 
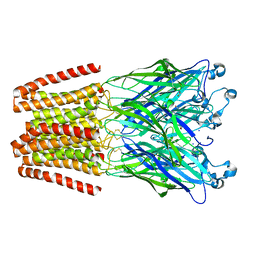 | | X-ray structure of the 2-22' locally-closed mutant of GLIC in complex with cyanoselenobarbital (seleniated barbiturate) | | Descriptor: | 2-[5-ethyl-2,4,6-tris(oxidanylidene)-1,3-diazinan-5-yl]ethyl selenocyanate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Reinholds Ruza, R, Fourati, Z, Delarue, M. | | Deposit date: | 2016-05-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Barbiturates Bind in the GLIC Ion Channel Pore and Cause Inhibition by Stabilizing a Closed State.
J. Biol. Chem., 292, 2017
|
|
7PJR
 
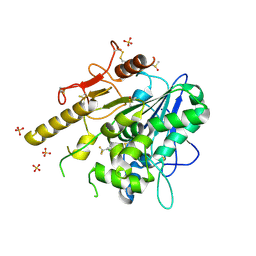 | | Notum_ARUK3000438 | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-1,2,3-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Zhao, Y, Fish, P, Jones, E.Y. | | Deposit date: | 2021-08-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Design of a Potent, Selective, and Brain-Penetrant Inhibitor of Wnt-Deactivating Enzyme Notum by Optimization of a Crystallographic Fragment Hit.
J.Med.Chem., 65, 2022
|
|
7PK3
 
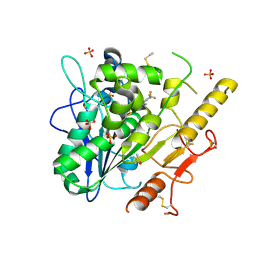 | | Notum_ARUK3001185 | | Descriptor: | 1-[2,4-bis(chloranyl)-3-(trifluoromethyl)phenyl]-1,2,3-triazole, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Vecchia, L, Hillier, J, Zhao, Y, Fish, P, Jones, E.Y. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Design of a Potent, Selective, and Brain-Penetrant Inhibitor of Wnt-Deactivating Enzyme Notum by Optimization of a Crystallographic Fragment Hit.
J.Med.Chem., 65, 2022
|
|
6R8P
 
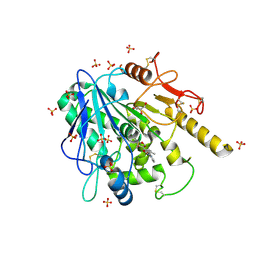 | | Notum fragment 723 | | Descriptor: | 2-(2-methylphenoxy)-~{N}-pyridin-3-yl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
5MUR
 
 | | X-ray structure of the F14'A mutant of GLIC in complex with propofol | | Descriptor: | 2,6-BIS(1-METHYLETHYL)PHENOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Fourati, Z, Delarue, M. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
5MVN
 
 | |
5MZQ
 
 | | X-ray structure of the M205W mutant of GLIC in complex with bromoform | | Descriptor: | (3R)-3-(dodecanoyloxy)tetradecanoic acid, 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, ... | | Authors: | Fourati, Z, Delarue, M. | | Deposit date: | 2017-02-01 | | Release date: | 2018-02-28 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
5NKJ
 
 | | X-ray structure of the N239C mutant of GLIC | | Descriptor: | Proton-gated ion channel | | Authors: | Hu, H, Fourati, Z, Delarue, M. | | Deposit date: | 2017-03-31 | | Release date: | 2018-06-20 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | Structural Basis for a Bimodal Allosteric Mechanism of General Anesthetic Modulation in Pentameric Ligand-Gated Ion Channels.
Cell Rep, 23, 2018
|
|
6ZVL
 
 | | ARUK3000263 complex with Notum | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[4-chloranyl-3-(trifluoromethyl)phenyl]-3~{H}-1,3,4-oxadiazol-2-one, ... | | Authors: | Zhao, Y, Ruza, R. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 5-Phenyl-1,3,4-oxadiazol-2(3 H )-ones Are Potent Inhibitors of Notum Carboxylesterase Activity Identified by the Optimization of a Crystallographic Fragment Screening Hit.
J.Med.Chem., 63, 2020
|
|
