1YGW
 
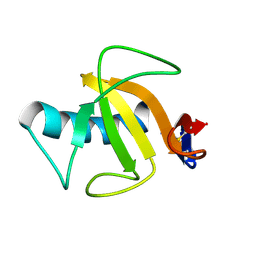 | |
3BYC
 
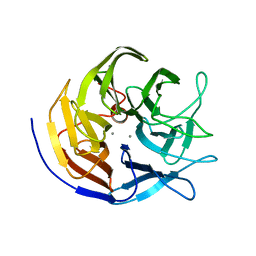 | | Joint neutron and X-ray structure of diisopropyl fluorophosphatase. Deuterium occupancies are 1-Q, where Q is occupancy of H | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Blum, M.-M, Mustyakimov, M, Ruterjans, H, Schoenborn, B.P, Langan, P, Chen, J.C.-H. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (2.2 Å), X-RAY DIFFRACTION | | Cite: | Rapid determination of hydrogen positions and protonation states of diisopropyl fluorophosphatase by joint neutron and X-ray diffraction refinement.
Proc.Natl.Acad.Sci.Usa, 106, 2009
|
|
2GVX
 
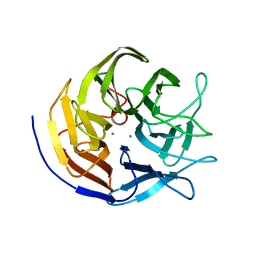 | | Structure of diisopropyl fluorophosphatase (DFPase), mutant D229N / N175D | | Descriptor: | CALCIUM ION, diisopropyl fluorophosphatase | | Authors: | Blum, M.-M, Lohr, F, Richardt, A, Ruterjans, H, Chen, J.C.-H. | | Deposit date: | 2006-05-03 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of a Designed Substrate Analogue to Diisopropyl Fluorophosphatase: Implications for the Phosphotriesterase Mechanism
J.Am.Chem.Soc., 128, 2006
|
|
1DF3
 
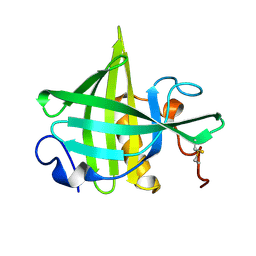 | | SOLUTION STRUCTURE OF A RECOMBINANT MOUSE MAJOR URINARY PROTEIN | | Descriptor: | MAJOR URINARY PROTEIN | | Authors: | Luecke, C, Franzoni, L, Abbate, F, Loehr, F, Ferrari, E, Sorbi, R.T, Rueterjans, H, Spisni, A. | | Deposit date: | 1999-11-17 | | Release date: | 2000-05-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant mouse major urinary protein.
Eur.J.Biochem., 266, 1999
|
|
1C7M
 
 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | HEME C, PROTEIN (CYTOCHROME C552) | | Authors: | Pristovsek, P, Luecke, C, Reincke, B, Ludwig, B, Rueterjans, H. | | Deposit date: | 2000-03-03 | | Release date: | 2000-07-19 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the functional domain of Paracoccus denitrificans cytochrome c552 in the reduced state.
Eur.J.Biochem., 267, 2000
|
|
3KGG
 
 | | X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase): Perdeuteration of proteins for neutron diffraction | | Descriptor: | CALCIUM ION, Diisopropyl-fluorophosphatase | | Authors: | Blum, M.-M, Tomanicek, S.J, John, H, Hanson, B.L, terjans, H.R, Schoenborn, B.P, Langan, P, Chen, J.C.-H. | | Deposit date: | 2009-10-29 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of perdeuterated diisopropyl fluorophosphatase (DFPase): perdeuteration of proteins for neutron diffraction.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
1BWY
 
 | | NMR STUDY OF BOVINE HEART FATTY ACID BINDING PROTEIN | | Descriptor: | PROTEIN (HEART FATTY ACID BINDING PROTEIN) | | Authors: | Lassen, D, Luecke, C, Kveder, M, Mesgarzadeh, A, Schmidt, J.M, Specht, B, Lezius, A, Spener, F, Rueterjans, H. | | Deposit date: | 1998-09-29 | | Release date: | 1998-10-07 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of bovine heart fatty-acid-binding protein with bound palmitic acid, determined by multidimensional NMR spectroscopy.
Eur.J.Biochem., 230, 1995
|
|
1EAL
 
 | | NMR STUDY OF ILEAL LIPID BINDING PROTEIN | | Descriptor: | ILEAL LIPID BINDING PROTEIN | | Authors: | Luecke, C, Zhang, F, Rueterjans, H, Hamilton, J.A, Sacchettini, J.C. | | Deposit date: | 1996-08-28 | | Release date: | 1997-01-22 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Flexibility is a likely determinant of binding specificity in the case of ileal lipid binding protein.
Structure, 4, 1996
|
|
1PM6
 
 | | Solution Structure of Full-Length Excisionase (Xis) from Bacteriophage HK022 | | Descriptor: | Excisionase | | Authors: | Rogov, V.V, Luecke, C, Muresanu, L, Wienk, H, Kleinhaus, I, Werner, K, Loehr, F, Pristovsek, P, Rueterjans, H. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-30 | | Last modified: | 2021-10-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure and stability of the full-length excisionase from bacteriophage HK022.
Eur.J.Biochem., 270, 2003
|
|
1PJX
 
 | | 0.85 ANGSTROM STRUCTURE OF SQUID GANGLION DFPASE | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, ... | | Authors: | Koepke, J, Rueterjans, H, Luecke, C, Fritzsch, G. | | Deposit date: | 2003-06-04 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Statistical analysis of crystallographic data obtained from squid ganglion DFPase at 0.85 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1I6E
 
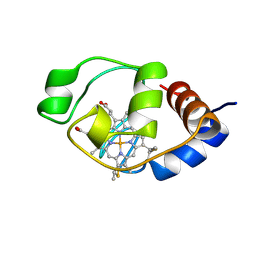 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE OXIDIZED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1P4W
 
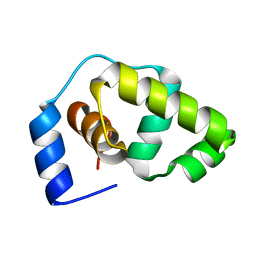 | | Solution structure of the DNA-binding domain of the Erwinia amylovora RcsB protein | | Descriptor: | rcsB | | Authors: | Pristovsek, P, Sengupta, K, Loehr, F, Schaefer, B, Wehland von Trebra, M, Rueterjans, H, Bernhard, F. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the DNA-binding domain of the Erwinia amylovora RcsB protein and its interaction with the RcsAB box.
J.Biol.Chem., 278, 2003
|
|
1G5W
 
 | | SOLUTION STRUCTURE OF HUMAN HEART-TYPE FATTY ACID BINDING PROTEIN | | Descriptor: | FATTY ACID-BINDING PROTEIN | | Authors: | Luecke, C, Rademacher, M, Zimmerman, A, van Moerkerk, H.T.B, Veerkamp, J.H, Rueterjans, H. | | Deposit date: | 2000-11-02 | | Release date: | 2001-03-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Spin-system heterogeneities indicate a selected-fit mechanism in fatty acid binding to heart-type fatty acid-binding protein (H-FABP).
Biochem.J., 354, 2001
|
|
1I6D
 
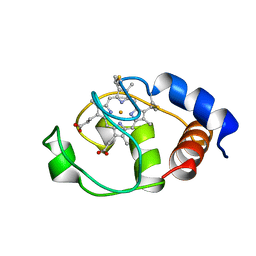 | | SOLUTION STRUCTURE OF THE FUNCTIONAL DOMAIN OF PARACOCCUS DENITRIFICANS CYTOCHROME C552 IN THE REDUCED STATE | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Reincke, B, Perez, C, Pristovsek, P, Luecke, C, Ludwig, C, Loehr, F, Rogov, V.V, Ludwig, B, Rueterjans, H. | | Deposit date: | 2001-03-02 | | Release date: | 2001-10-17 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the functional domain of Paracoccus denitrificans cytochrome c(552) in both redox states.
Biochemistry, 40, 2001
|
|
1L6U
 
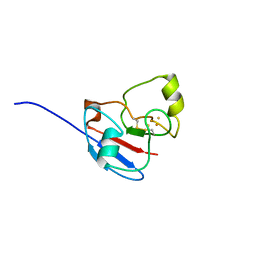 | | NMR STRUCTURE OF OXIDIZED ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
2GVU
 
 | |
2GVV
 
 | |
2GVW
 
 | |
1HXV
 
 | | PPIASE DOMAIN OF THE MYCOPLASMA GENITALIUM TRIGGER FACTOR | | Descriptor: | TRIGGER FACTOR | | Authors: | Vogtherr, M, Parac, T.N, Maurer, M, Pahl, A, Fiebig, K. | | Deposit date: | 2001-01-17 | | Release date: | 2002-05-29 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and dynamics of the peptidyl-prolyl cis-trans isomerase domain of the trigger factor from Mycoplasma genitalium compared to FK506-binding protein.
J.Mol.Biol., 318, 2002
|
|
1JBH
 
 | | Solution structure of cellular retinol binding protein type-I in the ligand-free state | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-06-04 | | Release date: | 2002-06-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of Apo- and holo-cellular retinol-binding protein in solution.
J.Biol.Chem., 277, 2002
|
|
1JJX
 
 | | Solution Structure of Recombinant Human Brain-type Fatty acid Binding Protein | | Descriptor: | BRAIN-TYPE FATTY ACID BINDING PROTEIN | | Authors: | Rademacher, M, Zimmerman, A.W, Rueterjans, H, Veerkamp, J.H, Luecke, C. | | Deposit date: | 2001-07-10 | | Release date: | 2002-10-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of fatty acid-binding protein from human brain.
Mol.Cell.Biochem., 239, 2002
|
|
1KGL
 
 | | Solution structure of cellular retinol binding protein type-I in complex with all-trans-retinol | | Descriptor: | CELLULAR RETINOL-BINDING PROTEIN TYPE I, RETINOL | | Authors: | Franzoni, L, Luecke, C, Perez, C, Cavazzini, D, Rademacher, M, Ludwig, C, Spisni, A, Rossi, G.L, Rueterjans, H. | | Deposit date: | 2001-11-27 | | Release date: | 2002-06-19 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure and Backbone Dynamics of Apo- and Holo-cellular Retinol-binding
Protein in Solution.
J.Biol.Chem., 277, 2002
|
|
1L6V
 
 | | STRUCTURE OF REDUCED BOVINE ADRENODOXIN | | Descriptor: | Adrenodoxin 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Beilke, D, Weiss, R, Lohr, F, Pristovsek, P, Hannemann, F, Bernhardt, R, Rueterjans, H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-06-26 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A new electron transport mechanism in mitochondrial steroid hydroxylase systems based on structural changes upon the reduction of adrenodoxin.
Biochemistry, 41, 2002
|
|
1LEB
 
 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
1LEA
 
 | | SOLUTION STRUCTURE OF THE LEXA REPRESSOR DNA BINDING DETERMINED BY 1H NMR SPECTROSCOPY | | Descriptor: | LEXA REPRESSOR DNA BINDING DOMAIN | | Authors: | Fogh, R.H, Ottleben, G, Rueterjans, H, Schnarr, M, Boelens, R, Kaptein, R. | | Deposit date: | 1994-05-11 | | Release date: | 1994-08-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy.
EMBO J., 13, 1994
|
|
