3ZFJ
 
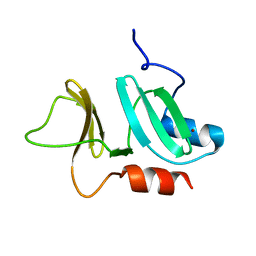 | | N-terminal domain of pneumococcal PhtD protein with bound Zn(II) | | Descriptor: | PNEUMOCOCCAL HISTIDINE TRIAD PROTEIN D, ZINC ION | | Authors: | Bersch, B, Bougault, C, Favier, A, Gabel, F, Roux, L, Vernet, T, Durmort, C. | | Deposit date: | 2012-12-11 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | New Insights Into Histidine Triad Proteins: Solution Structure of a Streptococcus Pneumoniae Phtd Domain and Zinc Transfer to Adcaii.
Plos One, 8, 2013
|
|
4ELC
 
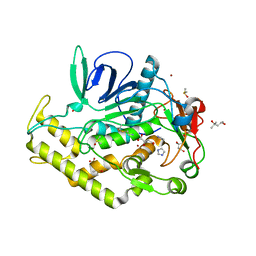 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134 mutant with MTSEA modified Cys-165 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Botulinum neurotoxin A light chain, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Bakirci, H, Garcia, S, Dive, V. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4EJ5
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A wild-type | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, CARBONATE ION, ... | | Authors: | Stura, E.A, Vera, L, Dive, V. | | Deposit date: | 2012-04-06 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4EL4
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S/C165S double mutant | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, GLYCEROL, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Dive, V. | | Deposit date: | 2012-04-10 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Framework for Covalent Inhibition of Clostridium botulinum Neurotoxin A by Targeting Cys165.
J.Biol.Chem., 287, 2012
|
|
4KS6
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S mutant with covalent inhibitor that modifies Cys-165 causing disorder in 166-174 stretch | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stura, E.A, Vera, L, Guitot, K, Dive, V. | | Deposit date: | 2013-05-17 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
4KUF
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134 mutant with MTSEA modified Cys-165 causing stretch disorder | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, Botulinum neurotoxin A light chain, ... | | Authors: | Stura, E.A, Vera, L, Ptchelkine, D, Bakirci, H, Garcia, S, Dive, V. | | Deposit date: | 2013-05-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.697 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
4KTX
 
 | | Crystal structure of the catalytic domain of botulinum neurotoxin BoNT/A C134S mutant with covalent inhibitor that modifies Cys-165 causing disorder in 167-174 stretch | | Descriptor: | Botulinum neurotoxin A light chain, GLYCEROL, Peptide inhibitor MPT-DPP-ARG-G-LEU-NH2, ... | | Authors: | Stura, E.A, Vera, L, Guitot, K, Dive, V. | | Deposit date: | 2013-05-21 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Covalent modification of the active site cysteine stresses Clostridium botulinum neurotoxin A
To be Published
|
|
4C6A
 
 | | High Resolution Structure of the Nucleoside diphosphate kinase | | Descriptor: | DI(HYDROXYETHYL)ETHER, NUCLEOSIDE DIPHOSPHATE KINASE, CYTOSOLIC | | Authors: | Priet, S, Ferron, F, Alvarez, K, Verron, M, Canard, B. | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Enzymatic Synthesis of Acyclic Nucleoside Thiophosphonate Diphosphates: Effect of the Alpha-Phosphorus Configuration on HIV-1 RT Activity.
Antiviral Res., 117, 2015
|
|
4CP5
 
 | | ndpK in complex with (Rp)-SPMPApp | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE, CYTOSOLIC, [[(2R)-1-(6-aminopurin-9-yl)propan-2-yl]oxymethyl-sulfanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Priet, S, Ferron, F, Alvarez, K, Verron, M, Canard, B. | | Deposit date: | 2014-01-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Enzymatic Synthesis of Acyclic Nucleoside Thiophosphonate Diphosphates: Effect of the Alpha-Phosphorus Configuration on HIV-1 RT Activity.
Antiviral Res., 117, 2015
|
|
5FVI
 
 | | Structure of IrisFP in mineral grease at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
5FVF
 
 | | Room temperature structure of IrisFP determined by serial femtosecond crystallography. | | Descriptor: | AMMONIUM ION, Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-06 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett., 7, 2016
|
|
5FVG
 
 | | Structure of IrisFP at 100 K. | | Descriptor: | Green to red photoconvertible GFP-like protein EosFP, SULFATE ION | | Authors: | Colletier, J.P, Gallat, F.X, Coquelle, N, Weik, M. | | Deposit date: | 2016-02-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Serial Femtosecond Crystallography and Ultrafast Absorption Spectroscopy of the Photoswitchable Fluorescent Protein Irisfp.
J.Phys.Chem.Lett, 7, 2016
|
|
