6MJV
 
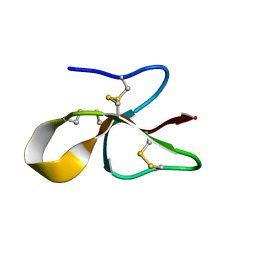 | | A consensus human beta defensin | | Descriptor: | Human beta-defensin | | Authors: | Amero, C, Villegas-Moreno, J, Rodriguez, A, Norton, R.S, Corzo, G. | | Deposit date: | 2018-09-22 | | Release date: | 2019-08-28 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antimicrobial activity and structure of a consensus human beta-defensin and its comparison to a novel putative hBD10.
Proteins, 88, 2020
|
|
8B2A
 
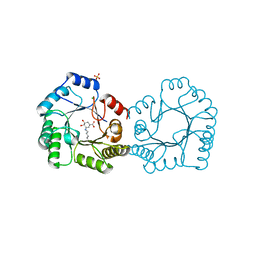 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2C
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (1~{S},2~{R},4~{R},5~{S},6~{S})-2,4,5-trihydroxy-7-oxabicyclo[4.1.0]heptane-2-carboxylic acid, 3-dehydroquinate dehydratase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2B
 
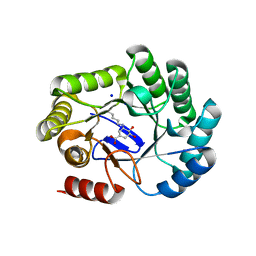 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, SODIUM ION | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
6Q84
 
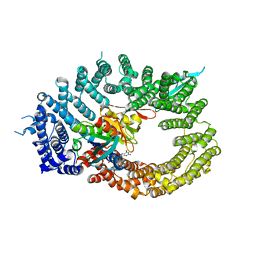 | | Crystal structure of RanGTP-Pdr6-eIF5A export complex | | Descriptor: | Eukaryotic translation initiation factor 5A-1, GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
6Q83
 
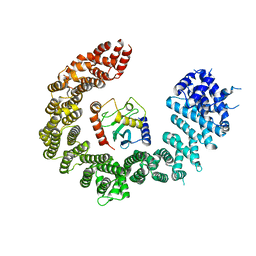 | | Crystal structure of the biportin Pdr6 in complex with UBC9 | | Descriptor: | Importin beta-like protein KAP122, UBC9 | | Authors: | Aksu, M, Trakhanov, S, Vera-Rodriguez, A, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (4.53 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
6Q82
 
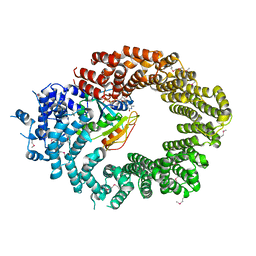 | | Crystal structure of the biportin Pdr6 in complex with RanGTP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-TRIPHOSPHATE, Importin beta-like protein KAP122, ... | | Authors: | Aksu, M, Vera-Rodriguez, A, Trakhanov, S, Gorlich, D. | | Deposit date: | 2018-12-14 | | Release date: | 2019-05-01 | | Last modified: | 2019-06-12 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural basis for the nuclear import and export functions of the biportin Pdr6/Kap122.
J.Cell Biol., 218, 2019
|
|
3LRL
 
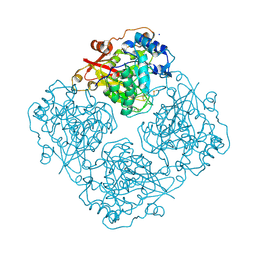 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae with melibiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3LRK
 
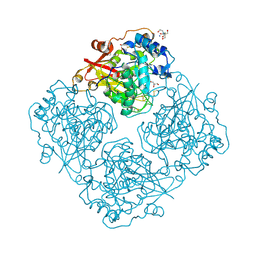 | | Structure of alfa-galactosidase (MEL1) from Saccharomyces cerevisiae | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3LRM
 
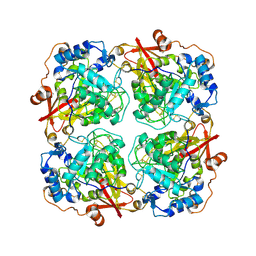 | | Structure of alfa-galactosidase from Saccharomyces cerevisiae with raffinose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Cerdan, M.E, Becerra, M, Sanz-Aparicio, J. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of Saccharomyces cerevisiae alpha-galactosidase and its complexes with natural substrates reveals new insights into substrate specificity of GH27 glycosidases.
J.Biol.Chem., 285, 2010
|
|
3OB8
 
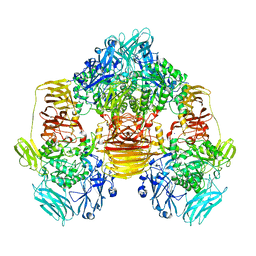 | | Structure of the beta-galactosidase from Kluyveromyces lactis in complex with galactose | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
3OBA
 
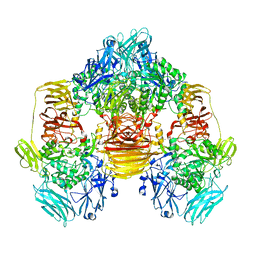 | | Structure of the beta-galactosidase from Kluyveromyces lactis | | Descriptor: | Beta-galactosidase, GLYCEROL, MANGANESE (III) ION | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
4C9F
 
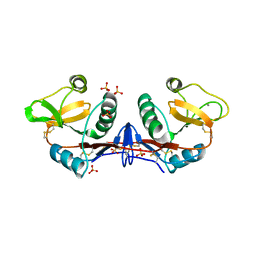 | | Structure of SIGN-R1 in complex with Sulfodextran | | Descriptor: | 4-O-sulfo-alpha-D-glucopyranose, CALCIUM ION, CD209 ANTIGEN-LIKE PROTEIN B, ... | | Authors: | Silva-Martin, N, Bartual, S.G, Rodriguez, A, Ramirez, E, Chacon, P, Anthony, R.M, Park, C.G, Hermoso, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
6FVB
 
 | |
5ARC
 
 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, GLYCEROL, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
5ARB
 
 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
5ARD
 
 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | 5-methyl-2-(5-methylpyridin-2-yl)pyridine, CALCIUM ION, GLYCEROL, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-24 | | Release date: | 2016-09-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cooperative Bio-Metallic Selectivity in a Tailored Protease Enables Creation of a C-C Cross-Coupling Heckase
To be Published
|
|
5AQE
 
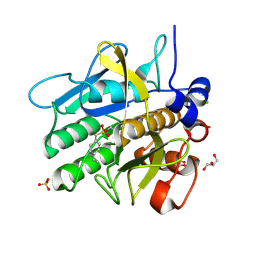 | | Cooperative bio-metallic selectivity in a tailored protease enables creation of a C-C cross-coupling Heckase | | Descriptor: | (4-VINYLPHENYL)METHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Sharma, M, Diaz-Rodriguez, A, Offen, W.A, Palm-Espling, M.E, Pordea, A, Wormald, M.R, Mcdonough, M, Davies, G.J, Davis, B.G. | | Deposit date: | 2015-09-22 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Bisubstrate Localisation by a Tailored Serine Protease Allows Creation of a Heck-Ase
To be Published
|
|
4LXJ
 
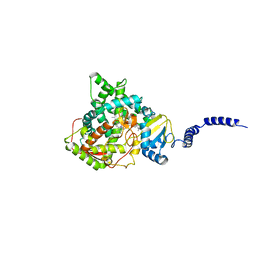 | | Saccharomyces cerevisiae lanosterol 14-alpha demethylase with lanosterol bound | | Descriptor: | LANOSTEROL, Lanosterol 14-alpha demethylase, OXYGEN MOLECULE, ... | | Authors: | Monk, B.C, Tomasiak, T.M, Keniya, M.V, Huschmann, F.U, Tyndall, J.D.A, O'Connell III, J.D, Cannon, R.D, McDonald, J, Rodriguez, A, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6TUK
 
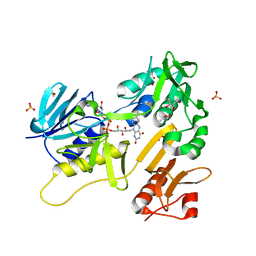 | | Crystal structure of Fdr9 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, Putative oxidoreductase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification and crystal structure determination of a ferredoxin reductase from the actinobacterium Thermobifida fusca.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6TO2
 
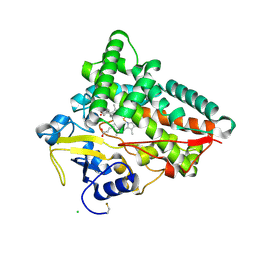 | | Crystal structure of CYP154C5 from Nocardia farcinica in complex with 5alpha-Androstan-3-one | | Descriptor: | (5~{S},8~{S},9~{S},10~{S},13~{S},14~{S})-10,13-dimethyl-1,2,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydrocyclopenta[a]phenanthren-3-one, CHLORIDE ION, Cytochrome P450 monooxygenase, ... | | Authors: | Rodriguez, A, Kluenemann, T, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CYP154C5 Regioselectivity in Steroid Hydroxylation Explored by Substrate Modifications and Protein Engineering*.
Chembiochem, 22, 2021
|
|
4R6Z
 
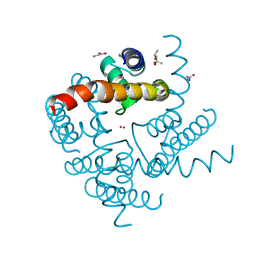 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, Cs+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CESIUM ION, GLYCINE, ... | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-08-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4R8C
 
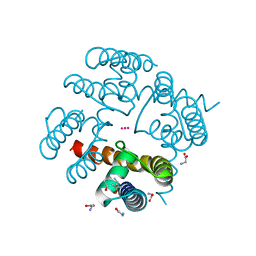 | | Crystal Structure of CNG mimicking NaK-ETPP mutant in complex with Rb+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, Potassium channel protein, ... | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-09-01 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RAI
 
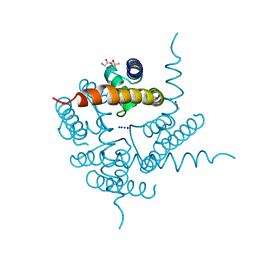 | | Crystal Structure of CNG mimicking NaK-ETPP mutant in complex with Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, Potassium channel protein, ... | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-09-10 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RO2
 
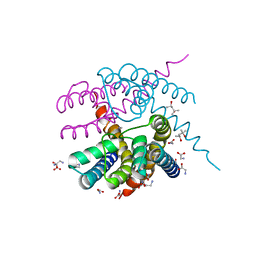 | | Crystal Structure of CNG mimicking NaK-ETPP mutant cocrystallized with Methylammonium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, Potassium channel protein, ... | | Authors: | De March, M, Napolitano, L.M.R, Onesti, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural, functional, and computational analysis suggests pore flexibility as the base for the poor selectivity of CNG channels.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
