6H9S
 
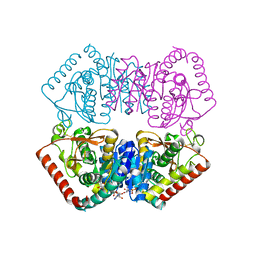 | |
2MK3
 
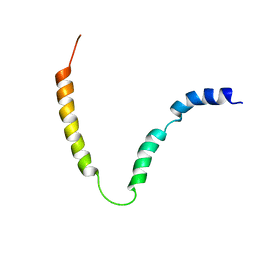 | | Solution NMR structure of gp41 ectodomain monomer on a DPC micelle | | Descriptor: | Transmembrane glycoprotein, chimeric construct | | Authors: | Roche, J, Louis, J.M, Grishaev, A, Ying, J, Bax, A. | | Deposit date: | 2014-01-23 | | Release date: | 2014-02-19 | | Last modified: | 2014-03-19 | | Method: | SOLUTION NMR | | Cite: | Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6QSS
 
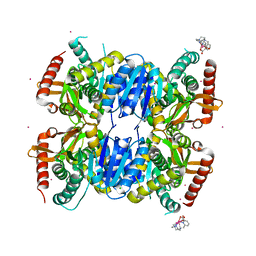 | | Crystal Structure of Ignicoccus islandicus malate dehydrogenase co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Roche, J, Girard, E, Madern, D. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (1.892 Å), X-RAY DIFFRACTION | | Cite: | The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.
J.Struct.Biol., 208, 2019
|
|
5LMW
 
 | | Llama nanobody PorM_02 | | Descriptor: | GLYCEROL, Nanobody | | Authors: | Roche, J, Gaubert, A, Leone, P, Roussel, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
6GZP
 
 | | Llama nanobody PorM_02 structure determined at room temperature by in-situ diffraction in ChipX microfluidic device | | Descriptor: | Nanobody | | Authors: | Roche, J, Gaubert, A, Desmyter, A, De Wijn, R, Sauter, C, Roussel, A. | | Deposit date: | 2018-07-04 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
7Q3X
 
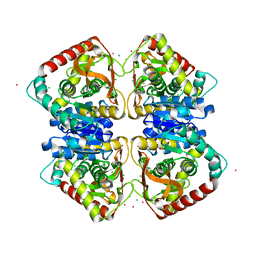 | |
6EY0
 
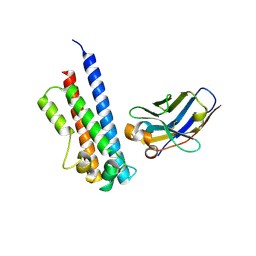 | | N-terminal part (residues 30-212) of PorM with the llama nanobody nb01 | | Descriptor: | T9SS component cytoplasmic membrane protein PorM, llama nanobody nb01 | | Authors: | Leone, P, Roche, J, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
2MJB
 
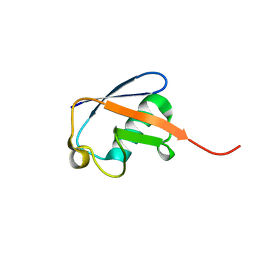 | | Solution nmr structure of ubiquitin refined against dipolar couplings in 4 media | | Descriptor: | Ubiquitin-60S ribosomal protein L40 | | Authors: | Maltsev, A, Grishaev, A, Roche, J, Zasloff, M, Bax, A. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Improved cross validation of a static ubiquitin structure derived from high precision residual dipolar couplings measured in a drug-based liquid crystalline phase.
J.Am.Chem.Soc., 136, 2014
|
|
6EY4
 
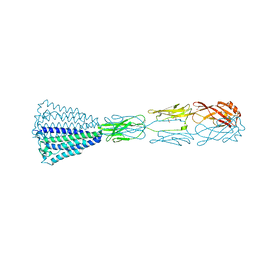 | | Periplasmic domain (residues 36-513) of GldM | | Descriptor: | 1,2-ETHANEDIOL, GldM | | Authors: | Leone, P, Roche, J, Cambillau, C, Roussel, A. | | Deposit date: | 2017-11-10 | | Release date: | 2018-02-07 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Type IX secretion system PorM and gliding machinery GldM form arches spanning the periplasmic space.
Nat Commun, 9, 2018
|
|
3MHB
 
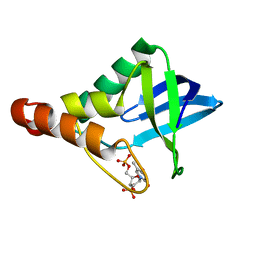 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L38A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-04-07 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MEH
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS I92A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-03-31 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MVV
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS F34A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-04 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MXP
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-07 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MZ5
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L103A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NK9
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V74A at cryogenic temperature | | Descriptor: | CALCIUM ION, Nuclease A, THYMIDINE-3',5'-DIPHOSPHATE | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2010-06-18 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NP8
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L36A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-06-28 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NQT
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V66A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-06-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NXW
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L125A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-07-14 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3OSO
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS L25A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3PMF
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Clark, I, Schlessman, J.L, Garcia-Moreno E, B, Heroux, A. | | Deposit date: | 2010-11-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3R3O
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS T62A at cryogenic temperature and with high redundancy | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno E, B. | | Deposit date: | 2011-03-16 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8EKP
 
 | | Apo rat TRPV2 in nanodiscs, state 1 | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.P, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular mechanisms and structural basis for TRPV2 acid-sensitivity
To Be Published
|
|
8EKQ
 
 | | Apo rat TRPV2 in nanodiscs, state 2 | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.P, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular mechanisms and structural basis for TRPV2 acid-sensitivity
To Be Published
|
|
8EKR
 
 | | Apo rat TRPV2 in nanodiscs, state 3 | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Pumroy, R.P, Moiseenkova-Bell, V.Y. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular mechanisms and structural basis for TRPV2 acid-sensitivity
To Be Published
|
|
8EKS
 
 | |
