5ML9
 
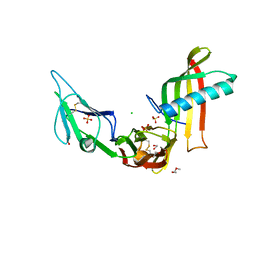 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer F4, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer F4 with specificity for Fc gamma receptor IIIa, CHLORIDE ION, ... | | Authors: | Robinson, J.I, Tomlinson, D.C, Baxter, E.W, Owen, R.L, Thomsen, M, Win, S.J, Nettleship, J.E, Tiede, C, Foster, R.J, Waterhouse, M.P, Harris, S.A, Owens, R.J, Fishwick, C.W.G, Goldman, A, McPherson, M.J, Morgan, A.W. | | Deposit date: | 2016-12-06 | | Release date: | 2017-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MN2
 
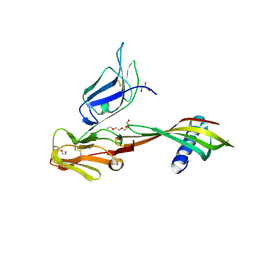 | | Cocrystal structure of Fc gamma receptor IIIa interacting with Affimer G3, a specific binding protein which blocks IgG binding to the receptor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Affimer G3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Robinson, J.I, Owen, R.L, Tomlinson, D.C, Baxter, E.W, Nettleship, J.E, Waterhouse, M.P, Harris, S.A, Owens, R.J, McPherson, M.J, Morgan, A.W, Tiede, C, Goldman, A, Thomsen, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-12-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Affimer proteins inhibit immune complex binding to Fc gamma RIIIa with high specificity through competitive and allosteric modes of action.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2LKY
 
 | |
2M0N
 
 | |
3NJB
 
 | |
3O2E
 
 | |
3OL3
 
 | |
3OL4
 
 | |
3OIB
 
 | |
3P96
 
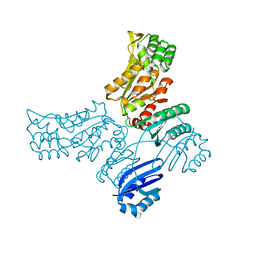 | |
3PFD
 
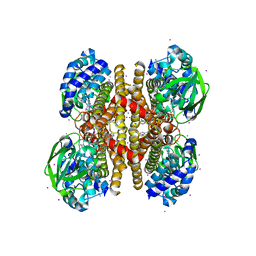 | |
3PM6
 
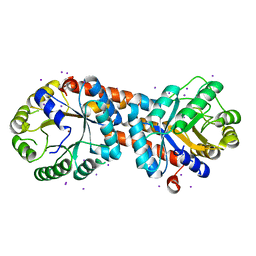 | |
3R2V
 
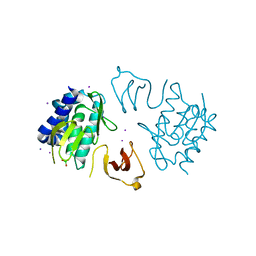 | |
3K9G
 
 | |
3KM3
 
 | |
3KW3
 
 | |
3LUZ
 
 | |
3MEN
 
 | |
5IS2
 
 | |
5IT4
 
 | |
5IT0
 
 | |
5JLR
 
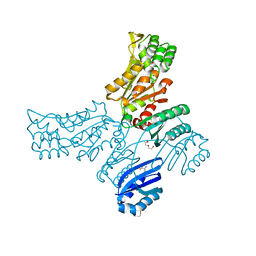 | | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, MAGNESIUM ION, ... | | Authors: | Shree, S, Agrawal, A, Dubey, S, Ramachandran, R. | | Deposit date: | 2016-04-27 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.261 Å) | | Cite: | Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain
To be published
|
|
5JJB
 
 | |
5JMA
 
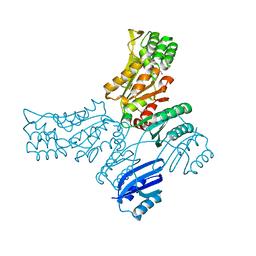 | | Crystal structure of Mycobacterium avium SerB2 in complex with serine at catalytic (PSP) domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Phosphoserine phosphatase, ... | | Authors: | Shree, S, Dubey, S, Agrawal, A, Ramachandran, R. | | Deposit date: | 2016-04-28 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Mycobacterium avium SerB2 in complex with serine at catalytic (PSP) domain
To be published
|
|
5JLP
 
 | |
