1RIN
 
 | | X-RAY CRYSTAL STRUCTURE OF A PEA LECTIN-TRIMANNOSIDE COMPLEX AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PEA LECTIN, ... | | Authors: | Rini, J.M, Hardman, K.D, Einspahr, H, Suddath, F.L, Carver, J.P. | | Deposit date: | 1993-01-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystal structure of a pea lectin-trimannoside complex at 2.6 A resolution.
J.Biol.Chem., 268, 1993
|
|
1HIN
 
 | |
1HIL
 
 | |
1GGI
 
 | | CRYSTAL STRUCTURE OF AN HIV-1 NEUTRALIZING ANTIBODY 50.1 IN COMPLEX WITH ITS V3 LOOP PEPTIDE ANTIGEN | | Descriptor: | HIV-1 V3 LOOP PEPTIDE ANTIGEN, IGG2A 50.1 FAB (HEAVY CHAIN), IGG2A 50.1 FAB (LIGHT CHAIN) | | Authors: | Stanfield, R.L, Rini, J.M, Wilson, I.A. | | Deposit date: | 1993-04-02 | | Release date: | 1993-10-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a human immunodeficiency virus type 1 neutralizing antibody, 50.1, in complex with its V3 loop peptide antigen.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
2AM4
 
 | |
2AM5
 
 | |
2APC
 
 | |
2AM3
 
 | |
1A3K
 
 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN GALECTIN-3 CARBOHYDRATE RECOGNITION DOMAIN (CRD) AT 2.1 ANGSTROM RESOLUTION | | Descriptor: | GALECTIN-3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seetharaman, J, Kanigsberg, A, Slaaby, R, Leffler, H, Barondes, S.H, Rini, J.M. | | Deposit date: | 1998-01-22 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-A resolution.
J.Biol.Chem., 273, 1998
|
|
5UB5
 
 | | human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus notch homolog protein 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
6ATK
 
 | |
3M2M
 
 | |
3OTK
 
 | | Structure and mechanisim of core 2 beta1,6-n-acetylglucosaminyltransferase: a Metal-ion independent gt-a glycosyltransferase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase, ... | | Authors: | Pak, J.E, Rini, J.M. | | Deposit date: | 2010-09-13 | | Release date: | 2011-09-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic characterization of leukocyte-type core 2 beta 1,6-N-acetylglucosaminyltransferase: a metal-ion-independent GT-A glycosyltransferase.
J.Mol.Biol., 414, 2011
|
|
3BGF
 
 | |
1C3D
 
 | | X-RAY CRYSTAL STRUCTURE OF C3D: A C3 FRAGMENT AND LIGAND FOR COMPLEMENT RECEPTOR 2 | | Descriptor: | C3D, GLYCEROL | | Authors: | Nagar, B, Jones, R.G, Diefenbach, R.J, Isenman, D.E, Rini, J.M. | | Deposit date: | 1998-05-19 | | Release date: | 1998-10-07 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of C3d: a C3 fragment and ligand for complement receptor 2.
Science, 280, 1998
|
|
5KY8
 
 | |
5L0R
 
 | | human POGLUT1 in complex with Notch1 EGF12 and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5KY7
 
 | | mouse POFUT1 in complex with O-glucosylated EGF(+) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EGF(+), GDP-fucose protein O-fucosyltransferase 1, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
2GAK
 
 | |
2GAM
 
 | | X-ray crystal structure of murine leukocyte-type Core 2 b1,6-N-acetylglucosaminyltransferase (C2GnT-L) in complex with Galb1,3GalNAc | | Descriptor: | beta-1,6-N-acetylglucosaminyltransferase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Pak, J.E, Rini, J.M. | | Deposit date: | 2006-03-09 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Crystal Structure of Leukocyte Type Core 2 beta1,6-N-Acetylglucosaminyltransferase: Evidence for a covergence of metal ion independent glycosyltransferase mechanism.
J.Biol.Chem., 281, 2006
|
|
6U7H
 
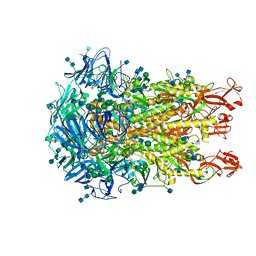 | | Cryo-EM structure of the HCoV-229E spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, Z, Benlekbir, S, Rubinstein, J.L, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6U7E
 
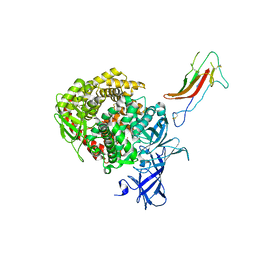 | | HCoV-229E RBD Class III in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6U7G
 
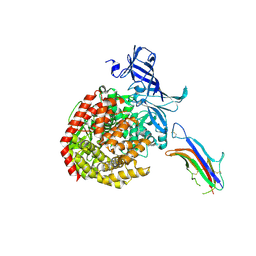 | | HCoV-229E RBD Class V in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
6U7F
 
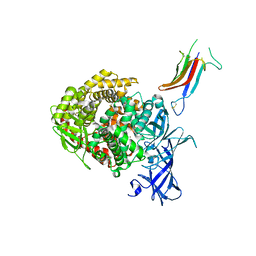 | | HCoV-229E RBD Class IV in complex with human APN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aminopeptidase N, ... | | Authors: | Tomlinson, A.C.A, Li, Z, Rini, J.M. | | Deposit date: | 2019-09-02 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
4FYQ
 
 | | Human aminopeptidase N (CD13) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Wong, A.H, Rini, J.M. | | Deposit date: | 2012-07-05 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-ray Crystal Structure of Human Aminopeptidase N Reveals a Novel Dimer and the Basis for Peptide Processing.
J.Biol.Chem., 287, 2012
|
|
