6HV2
 
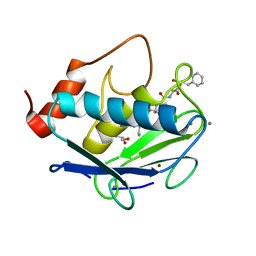 | | MMP-13 in complex with the peptide IMISF | | Descriptor: | CALCIUM ION, Collagenase 3, GLYCEROL, ... | | Authors: | Mittl, P, Riedl, R, Hohl, D. | | Deposit date: | 2018-10-10 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Drug Design Inspired by Nature: Crystallographic Detection of an Auto-Tailored Protease Inhibitor Template.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
4WV7
 
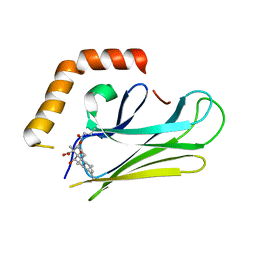 | | HEAT SHOCK PROTEIN 70 SUBSTRATE BINDING DOMAIN WITH COVALENTLY LINKED NOVOLACTONE | | Descriptor: | (5beta,6alpha,8alpha,14alpha)-13-ethenyl-5,6-dihydroxy-14-methylpodocarp-12-en-15-oic acid, Heat shock 70 kDa protein 1A/1B | | Authors: | Kirby, C.A, Baird, J, Stams, T. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The novolactone natural product disrupts the allosteric regulation of hsp70.
Chem.Biol., 22, 2015
|
|
4WV5
 
 | |
6XYE
 
 | |
6F0E
 
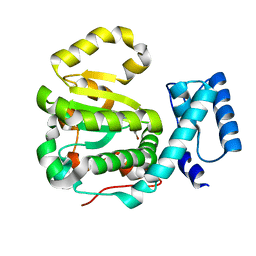 | | Structure of yeast Sec14p with a picolinamide compound | | Descriptor: | SEC14 cytosolic factor, ~{N}-(1,3-benzodioxol-5-ylmethyl)-5-bromanyl-3-fluoranyl-pyridine-2-carboxamide | | Authors: | Hong, Z, Johnen, P, Schaaf, G, Bono, F. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Target Identification and Mechanism of Action of Picolinamide and Benzamide Chemotypes with Antifungal Properties.
Cell Chem Biol, 25, 2018
|
|
2HD5
 
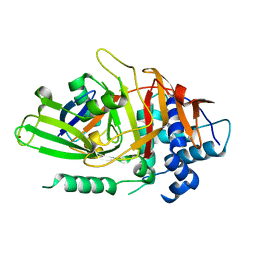 | | USP2 in complex with ubiquitin | | Descriptor: | Polyubiquitin, Ubiquitin carboxyl-terminal hydrolase 2, ZINC ION | | Authors: | Renatus, M, Kroemer, M. | | Deposit date: | 2006-06-20 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Ubiquitin Recognition by the Deubiquitinating Protease USP2.
Structure, 14, 2006
|
|
4FN5
 
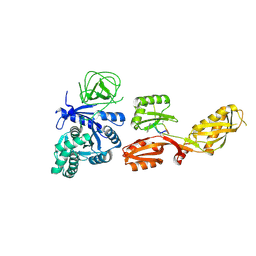 | |
5HL7
 
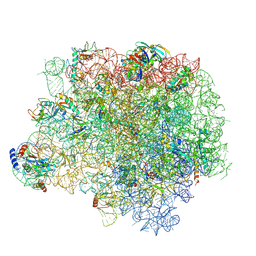 | | The crystal structure of the large ribosomal subunit of Staphylococcus aureus in complex with lefamulin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 23S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Eyal, Z, Matzov, D, Krupkin, M, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | A novel pleuromutilin antibacterial compound, its binding mode and selectivity mechanism.
Sci Rep, 6, 2016
|
|
6R0H
 
 | | Glycogen Phosphorylase b in complex with 3 | | Descriptor: | 3-(4-fluorophenyl)-~{N}-[(2~{R},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]benzamide, Glycogen phosphorylase, muscle form, ... | | Authors: | Tsagkarakou, S.A, Koulas, M.S, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-10 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
6R0I
 
 | | Glycogen Phosphorylase b in complex with 4 | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Koulas, M.S, Tsagkarakou, S.A, Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High Consistency of Structure-Based Design and X-Ray Crystallography: Design, Synthesis, Kinetic Evaluation and Crystallographic Binding Mode Determination of Biphenyl-N-acyl-beta-d-Glucopyranosylamines as Glycogen Phosphorylase Inhibitors.
Molecules, 24, 2019
|
|
7ZNY
 
 | | Cryo-EM structure of the canine distemper virus tetrameric attachment glycoprotein | | Descriptor: | Hemagglutinin glycoprotein | | Authors: | Kalbermatter, D, Jeckelmann, J.-M, Wyss, M, Plattet, P, Fotiadis, D. | | Deposit date: | 2022-04-23 | | Release date: | 2023-02-08 | | Last modified: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and supramolecular organization of the canine distemper virus attachment glycoprotein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8A1A
 
 | | Structure of a leucinostatin derivative determined by host lattice display : L1F11V1 construct | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, ... | | Authors: | Mittl, P.R.E. | | Deposit date: | 2022-06-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8A19
 
 | | Structure of a leucinostatin derivative determined by host lattice display : L1E4V1 construct | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-(2-methoxyethoxy)-11,15-dimethyl-8-oxa-2,11,15,19,21,23-hexazatetracyclo[15.6.1.13,7.020,24]pentacosa-1(23),3(25),4,6,17,20(24),21-heptaen-10-one, ... | | Authors: | Mittl, P.R.E. | | Deposit date: | 2022-06-01 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.358 Å) | | Cite: | Structure of a hydrophobic leucinostatin derivative determined by host lattice display.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
