4Q5W
 
 | | Crystal structure of extended-Tudor 9 of Drosophila melanogaster | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Maternal protein tudor | | Authors: | Ren, R, Liu, H, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-17 | | Release date: | 2014-05-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
4Q5Y
 
 | | Crystal structure of extended-Tudor 10-11 of Drosophila melanogaster | | Descriptor: | Maternal protein tudor | | Authors: | Liu, H, Ren, R, Wang, W, Wang, M, Yang, N, Dong, Y, Gong, W, Lehmann, R, Xu, R.M. | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and domain organization of Drosophila Tudor
Cell Res., 24, 2014
|
|
6WMI
 
 | | ZNF410 zinc fingers 1-5 with 17 mer blunt DNA Oligonucleotide | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*CP*AP*TP*AP*AP*TP*AP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*TP*AP*TP*GP*GP*GP*AP*TP*GP*TP*G)-3'), ... | | Authors: | Ren, R, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | ZNF410 Uniquely Activates the NuRD Component CHD4 to Silence Fetal Hemoglobin Expression.
Mol.Cell, 81, 2021
|
|
7N5U
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 21) | | Descriptor: | DNA Strain II, DNA Strand I, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5V
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 20) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
7N5W
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing GGACCC (Oligo 23) | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand I, DNA Strand II, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
8E3E
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#10) | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*A)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
8E3D
 
 | | ZBTB7A Zinc Finger Domain Bound to DNA Duplex Containing CAST sequence (#11) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*TP*TP*TP*GP*GP*GP*GP*AP*GP*GP*GP*GP*TP*CP*TP*TP*TP*AP*AP*CP*C)-3'), DNA (5'-D(P*GP*GP*TP*AP*AP*AP*AP*GP*AP*CP*CP*CP*CP*TP*CP*CP*CP*CP*AP*AP*AP*T)-3'), ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis for transcription factor ZBTB7A recognition of DNA and effects of ZBTB7A somatic mutations that occur in human acute myeloid leukemia.
J.Biol.Chem., 299, 2023
|
|
6ML5
 
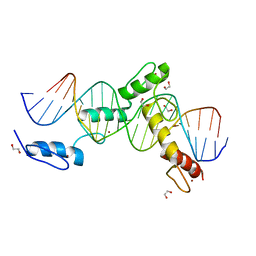 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML6
 
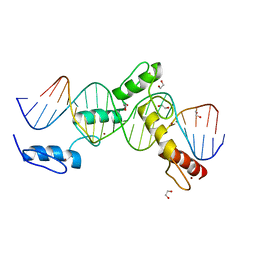 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpA 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*(5CM)P*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML4
 
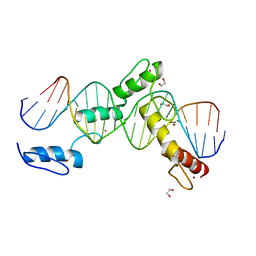 | | BTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 3) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*CP*GP*AP*AP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*GP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML2
 
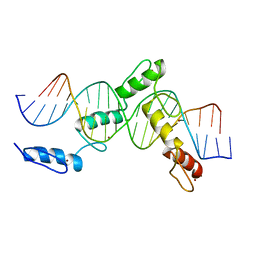 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 1) | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*CP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ZINC ION, ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.874 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML7
 
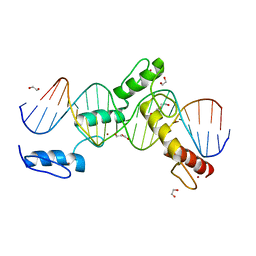 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 4 with a CpG 5mC Modification) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*(5CM)P*GP*AP*AP*TP*T)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
6ML3
 
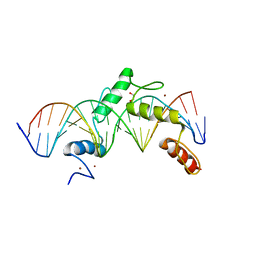 | | ZBTB24 Zinc Fingers 4-8 with 19+1mer DNA Oligonucleotide (Sequence 2) | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*CP*GP*CP*AP*GP*GP*TP*CP*CP*TP*GP*GP*AP*AP*GP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*CP*TP*TP*CP*CP*AP*GP*GP*AP*CP*CP*TP*GP*CP*G)-3'), ... | | Authors: | Horton, J.R, Cheng, X, Ren, R. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Structural basis of specific DNA binding by the transcription factor ZBTB24.
Nucleic Acids Res., 47, 2019
|
|
7N5T
 
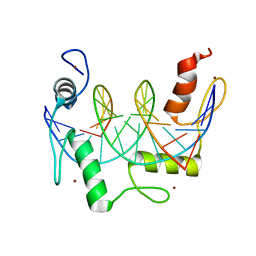 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 5) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7N5S
 
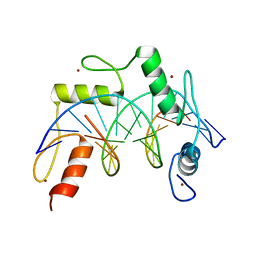 | | ZBTB7A Zinc Finger Domain Bound to -200 Site of Fetal Globin Promoter (Oligo 6) | | Descriptor: | DNA Strand I, DNA Strand II, ZINC ION, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2021-06-06 | | Release date: | 2021-08-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural basis for human ZBTB7A action at the fetal globin promoter.
Cell Rep, 36, 2021
|
|
7TXC
 
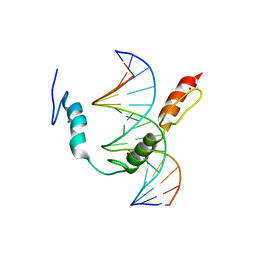 | | HIC2 zinc finger domain in complex with the DNA binding motif-2 of the BCL11A enhancer | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*TP*GP*GP*CP*AP*TP*TP*AP*TP*CP*T)-3'), DNA (5'-D(*AP*GP*AP*TP*AP*AP*TP*GP*CP*CP*AP*AP*CP*AP*GP*T)-3'), Hypermethylated in cancer 2 protein, ... | | Authors: | Horton, J.R, Ren, R, Cheng, X. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-01 | | Last modified: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | HIC2 controls developmental hemoglobin switching by repressing BCL11A transcription.
Nat.Genet., 54, 2022
|
|
4KUL
 
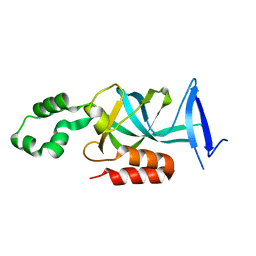 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUI
 
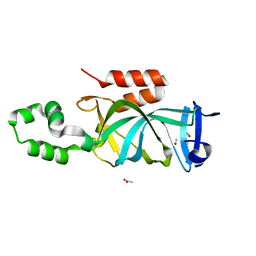 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3UDB
 
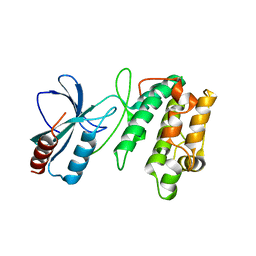 | | Crystal structure of SnRK2.6 | | Descriptor: | CHLORIDE ION, Serine/threonine-protein kinase SRK2E | | Authors: | Xie, T, Ren, R, Pang, Y, Yan, C. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.567 Å) | | Cite: | Molecular mechanism for the inhibition of a critical component in the Arabidopsis thaliana abscisic acid signal transduction pathways, SnRK2.6, by the protein phosphatase ABI1
to be published
|
|
4XU5
 
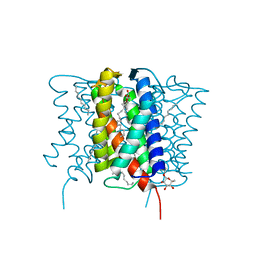 | | Crystal structure of MvINS bound to a bromine-derived 14C Diacylglycerol (DAG) at 2.1A resolution | | Descriptor: | (2S)-1-[(13-bromotridecanoyl)oxy]-3-hydroxypropan-2-yl tetradecanoate, DECANE, Uncharacterized protein, ... | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU4
 
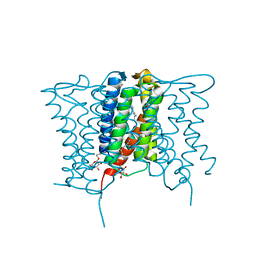 | | Crystal structure of a mycobacterial Insig homolog MvINS from Mycobacterium vanbaalenii at 1.9A resolution | | Descriptor: | DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Uncharacterized protein, nonyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
4XU6
 
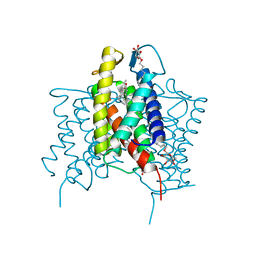 | | Crystal structure of cross-linked MvINS R77C trimer at 1.9A resolution | | Descriptor: | N-TRIDECANOIC ACID, Uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Ren, R.B, Wu, J.P, Yan, C.Y, He, Y, Yan, N. | | Deposit date: | 2015-01-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | PROTEIN STRUCTURE. Crystal structure of a mycobacterial Insig homolog provides insight into how these sensors monitor sterol levels
Science, 349, 2015
|
|
1ABO
 
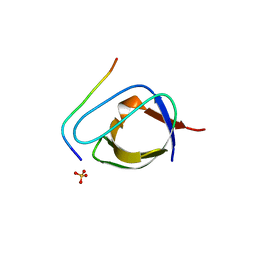 | | CRYSTAL STRUCTURE OF THE COMPLEX OF THE ABL TYROSINE KINASE SH3 DOMAIN WITH 3BP-1 SYNTHETIC PEPTIDE | | Descriptor: | 3BP-1 SYNTHETIC PEPTIDE, 10 RESIDUES, ABL TYROSINE KINASE, ... | | Authors: | Musacchio, A, Wilmanns, M, Saraste, M. | | Deposit date: | 1995-05-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides.
Nat.Struct.Biol., 1, 1994
|
|
