4GU3
 
 | |
4GU4
 
 | |
3TW5
 
 | | Crystal structure of the GP42 transglutaminase from Phytophthora sojae | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Transglutaminase elicitor | | Authors: | Reiss, K, Kirchner, E, Zocher, G, Stehle, T. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-12 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and Phylogenetic Analyses of the GP42 Transglutaminase from Phytophthora sojae Reveal an Evolutionary Relationship between Oomycetes and Marine Vibrio Bacteria.
J.Biol.Chem., 286, 2011
|
|
4XC5
 
 | | CRYSTAL STRUCTURE OF THE T1L REOVIRUS ATTACHMENT PROTEIN SIGMA1 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Reiss, K, Stehle, T. | | Deposit date: | 2014-12-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Serotype 1 Reovirus Attachment Protein sigma 1 in Complex with Junctional Adhesion Molecule A Reveals a Conserved Serotype-Independent Binding Epitope.
J.Virol., 89, 2015
|
|
6DW3
 
 | | SAMHD1 Bound to Cytarabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 4-amino-1-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}pyrimidin-2(1H)-one, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DW7
 
 | | SAMHD1 without Catalytic Nucleotides | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GLYCINE, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DW5
 
 | | SAMHD1 Bound to Gemcitabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-deoxy-2',2'-difluorocytidine 5'-(tetrahydrogen triphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DWJ
 
 | | SAMHD1 Bound to Vidarabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 9-{5-O-[(S)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-9H-purin-6-amine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DWD
 
 | | SAMHD1 Bound to Clofarabine-TP in the Catalytic Pocket and Allosteric Pocket | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, 9-{2-deoxy-2-fluoro-5-O-[(R)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-2-me thyl-9H-purin-6-amine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DW4
 
 | | SAMHD1 Bound to Cladribine-TP in the Catalytic Pocket and Allosteric Pocket | | Descriptor: | 2'-deoxy-2-methyladenosine 5'-(tetrahydrogen triphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DWK
 
 | | SAMHD1 Bound to Fludarabine-TP in the Catalytic Pocket | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-fluoro-9-{5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-9H-purin-6-a mine, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4EUK
 
 | | Crystal structure | | Descriptor: | 1,2-ETHANEDIOL, Histidine kinase 5, Histidine-containing phosphotransfer protein 1, ... | | Authors: | Stehle, T, Bauer, J. | | Deposit date: | 2012-04-25 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Function Analysis of Arabidopsis thaliana Histidine Kinase AHK5 Bound to Its Cognate Phosphotransfer Protein AHP1.
Mol Plant, 6, 2013
|
|
5MHR
 
 | | T3D reovirus sigma1 complexed with 9BG5 Fab fragments | | Descriptor: | 9BG5 Fab heavy chain, 9BG5 Fab light chain,LOC100046793 protein,MAb 110 light chain, Viral attachment protein sigma 1 | | Authors: | Stehle, T, Dietrich, M.H. | | Deposit date: | 2016-11-25 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insights into Reovirus sigma 1 Interactions with Two Neutralizing Antibodies.
J. Virol., 91, 2017
|
|
5MHS
 
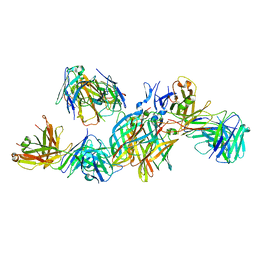 | | T1L reovirus sigma1 complexed with 5C6 Fab fragments | | Descriptor: | 5C6 Fab heavy chain, 5C6 Fab light chain, Outer capsid protein sigma-1 | | Authors: | Stehle, T, Dietrich, M.H. | | Deposit date: | 2016-11-25 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Insights into Reovirus sigma 1 Interactions with Two Neutralizing Antibodies.
J. Virol., 91, 2017
|
|
4ODB
 
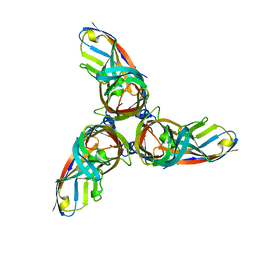 | |
6OYB
 
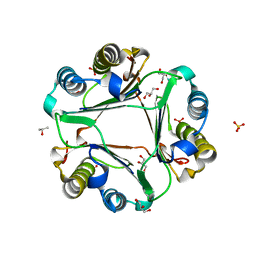 | |
6OYE
 
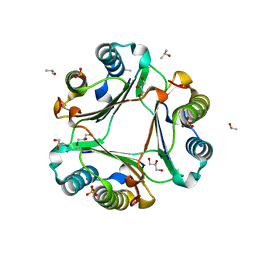 | |
6OYG
 
 | |
6OY8
 
 | |
7MRV
 
 | | F100A mutant structure of MIF2 (D-DT) | | Descriptor: | D-dopachrome decarboxylase, SULFATE ION | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MRU
 
 | | Crystal structure of S62A MIF2 mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, D-dopachrome decarboxylase | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-09 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MW7
 
 | | Crystal structure of P1G mutant of D-dopachrome tautomerase | | Descriptor: | D-dopachrome decarboxylase, SODIUM ION, SULFATE ION | | Authors: | Manjula, R, Murphy, E.L, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-15 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
7MSE
 
 | | High-resolution crystal structure of hMIF2 with tartrate at the active site | | Descriptor: | D-dopachrome decarboxylase, L(+)-TARTARIC ACID | | Authors: | Murphy, E.L, Manjula, R, Murphy, J.W, Lolis, E. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | A structurally preserved allosteric site in the MIF superfamily affects enzymatic activity and CD74 activation in D-dopachrome tautomerase.
J.Biol.Chem., 297, 2021
|
|
5UMK
 
 | |
5UMJ
 
 | |
