1XNF
 
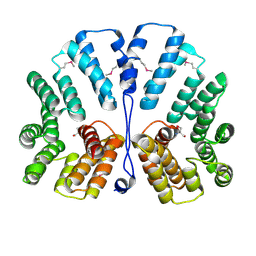 | | Crystal structure of E.coli TPR-protein NlpI | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Lipoprotein nlpI | | Authors: | Wilson, C.G, Kajander, T, Regan, L. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The crystal structure of NlpI. A prokaryotic tetratricopeptide repeat protein with a globular fold.
FEBS J., 272, 2005
|
|
1NA3
 
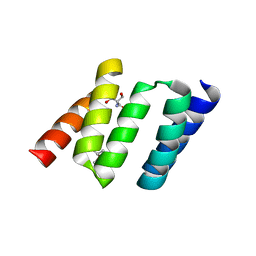 | | Design of Stable alpha-Helical Arrays from an Idealized TPR Motif | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Main, E, Xiong, Y, Cocco, M, D'Andrea, L, Regan, L. | | Deposit date: | 2002-11-26 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of Stable alpha-Helical Arrays from an Idealized TPR Motif
Structure, 11, 2003
|
|
1NA0
 
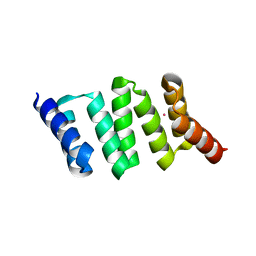 | | Design of Stable alpha-Helical Arrays from an Idealized TPR Motif | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, CHLORIDE ION, ... | | Authors: | Main, E, Xiong, Y, Cocco, M, D'Andrea, L, Regan, L. | | Deposit date: | 2002-11-26 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of Stable alpha-Helical Arrays from an Idealized TPR Motif
Structure, 11, 2003
|
|
3ESK
 
 | |
2AVP
 
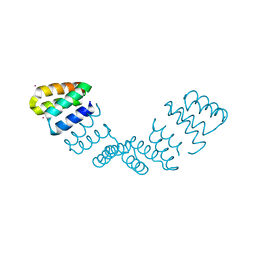 | | Crystal structure of an 8 repeat consensus TPR superhelix | | Descriptor: | CADMIUM ION, synthetic consensus TPR protein | | Authors: | Kajander, T, Cortajarena, A.L, Main, E.R, Mochrie, S, Regan, L. | | Deposit date: | 2005-08-30 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure and stability of designed TPR protein superhelices: unusual crystal packing and implications for natural TPR proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1F4M
 
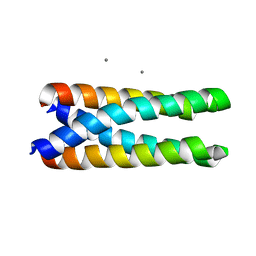 | | P3(2) CRYSTAL STRUCTURE OF ALA2ILE2-6, A VERSION OF ROP WITH A REPACKED HYDROPHOBIC CORE AND A NEW FOLD. | | Descriptor: | CALCIUM ION, ROP ALA2ILE2-6 | | Authors: | Willis, M.A, Bishop, B, Regan, L, Brunger, A.T. | | Deposit date: | 2000-06-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Dramatic structural and thermodynamic consequences of repacking a protein's hydrophobic core.
Structure Fold.Des., 8, 2000
|
|
1F4N
 
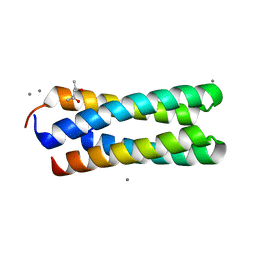 | | C2 CRYSTAL STRUCTURE OF ALA2ILE2-6, A VERSION OF ROP WITH A REPACKED HYDROPHOBIC CORE AND A NEW FOLD. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ROP ALA2ILE2-6 | | Authors: | Willis, M.A, Bishop, B, Regan, L, Brunger, A.T. | | Deposit date: | 2000-06-08 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dramatic structural and thermodynamic consequences of repacking a protein's hydrophobic core.
Structure Fold.Des., 8, 2000
|
|
2FO7
 
 | |
2HYZ
 
 | |
2QND
 
 | |
6CIU
 
 | | Structure of a Thr-rich interface in an Azami Green tetramer | | Descriptor: | Azami-Green | | Authors: | Oi, C, Lim, C.S, Knecht, K.M, Xiong, Y, Regan, L. | | Deposit date: | 2018-02-25 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A threonine zipper that mediates protein-protein interactions: Structure and prediction.
Protein Sci., 27, 2018
|
|
3FWV
 
 | | Crystal Structure of a Redesigned TPR Protein, T-MOD(VMY), in Complex with MEEVF Peptide | | Descriptor: | Heat shock protein HSP 90-beta, Hsc70/Hsp90-organizing protein, NICKEL (II) ION | | Authors: | Jackrel, M.E, Valverde, R, Regan, L. | | Deposit date: | 2009-01-19 | | Release date: | 2009-04-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Redesign of a protein-peptide interaction: characterization and applications
Protein Sci., 18, 2009
|
|
3KD7
 
 | |
1GTO
 
 | |
5DE2
 
 | |
6I2I
 
 | | Refined 13pf Hela Cell Tubulin microtubule (EML4-NTD decorated) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Atherton, J.M, Moores, C.A. | | Deposit date: | 2018-11-01 | | Release date: | 2019-08-28 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mitotic phosphorylation by NEK6 and NEK7 reduces the microtubule affinity of EML4 to promote chromosome congression.
Sci.Signal., 12, 2019
|
|
2WQO
 
 | | STRUCTURE OF NEK2 BOUND TO THE AMINOPYRIDINE CCT241950 | | Descriptor: | 4-[2-AMINO-5-(3,4,5-TRIMETHOXYPHENYL)PYRIDIN-3-YL]BENZOIC ACID, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK2 | | Authors: | Mas-Droux, C, Bayliss, R. | | Deposit date: | 2009-08-24 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.167 Å) | | Cite: | An Autoinhibitory Tyrosine Motif in the Cell-Cycle- Regulated Nek7 Kinase is Released Through Binding of Nek9.
Mol.Cell, 36, 2009
|
|
2WQM
 
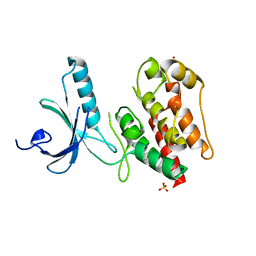 | | Structure of apo human Nek7 | | Descriptor: | NICKEL (II) ION, SERINE/THREONINE-PROTEIN KINASE NEK7, SULFATE ION | | Authors: | Richards, M.W, Bayliss, R. | | Deposit date: | 2009-08-24 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Auto-Inhibitory Tyrosine Motif in the Cell-Cycle Regulated Nek7 Kinase is Released Through Binding of Nek9
Mol.Cell, 36, 2009
|
|
2WQN
 
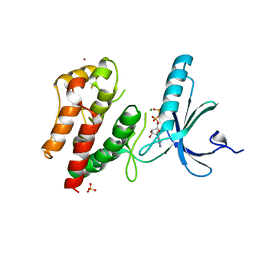 | | Structure of ADP-bound human Nek7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Richards, M.W, Bayliss, R. | | Deposit date: | 2009-08-24 | | Release date: | 2009-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | An Autoinhibitory Tyrosine Motif in the Cell-Cycle- Regulated Nek7 Kinase is Released Through Binding of Nek9.
Mol.Cell, 36, 2009
|
|
6FHK
 
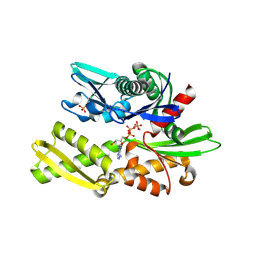 | |
4CGC
 
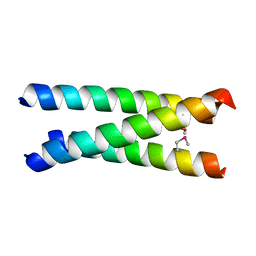 | |
4CI8
 
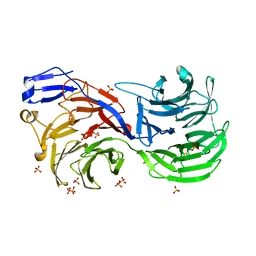 | |
4CGB
 
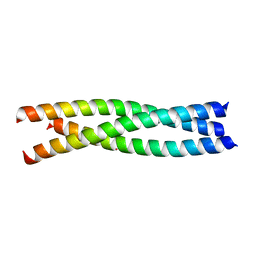 | | Crystal structure of the trimerization domain of EML2 | | Descriptor: | ECHINODERM MICROTUBULE-ASSOCIATED PROTEIN-LIKE 2, GLYCEROL, POTASSIUM ION | | Authors: | Richards, M.W, Bayliss, R. | | Deposit date: | 2013-11-21 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Microtubule Association of Eml Proteins and the Eml4-Alk Variant 3 Oncoprotein Require an N-Terminal Trimerization Domain.
Biochem.J., 467, 2015
|
|
4NDY
 
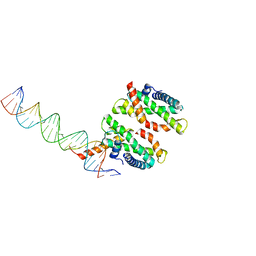 | | Human MHF1-MHF2 DNA complex | | Descriptor: | Centromere protein S, Centromere protein X, DNA (26-MER) | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.999 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
4NE1
 
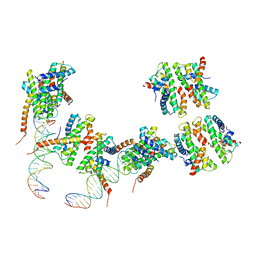 | | Human MHF1 MHF2 DNA complexes | | Descriptor: | Centromere protein S, Centromere protein X, DNA (26-MER) | | Authors: | Zhao, Q, Saro, D, Sachpatzidis, A, Sung, P, Xiong, Y. | | Deposit date: | 2013-10-28 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.499 Å) | | Cite: | The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes.
Nat Commun, 5, 2014
|
|
