8E5V
 
 | |
8E6I
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter M230A mutant in an inward-open, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E6H
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter A47W mutant in an occluded, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E6L
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter D296A mutant in an inward-open, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Divalent metal cation transporter MntH, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E60
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter WT in an occluded, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
8E5S
 
 | |
8E6M
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter WT in an inward-open, cadmium-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Ray, S, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
5KBG
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH OCRESOL | | Descriptor: | MopR, ZINC ION, o-cresol | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBI
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH CATACHOL | | Descriptor: | CATECHOL, MopR, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBE
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH PHENOL | | Descriptor: | MopR, PHENOL, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5KBH
 
 | | CRYSTAL STRUCTURE OF THE AROMATIC SENSOR DOMAIN OF MOPR IN COMPLEX WITH 3-CHLORO-PHENOL | | Descriptor: | 3-CHLOROPHENOL, MopR, ZINC ION | | Authors: | Ray, S, Anand, R, Panjikar, S. | | Deposit date: | 2016-06-03 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Basis of Selective Aromatic Pollutant Sensing by the Effector Binding Domain of MopR, an NtrC Family Transcriptional Regulator.
Acs Chem.Biol., 11, 2016
|
|
5XAZ
 
 | |
5XAY
 
 | |
8E6N
 
 | | X-ray structure of the Deinococcus radiodurans Nramp/MntH divalent transition metal transporter G223W mutant in an outward-open, manganese-bound state | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, DI(HYDROXYETHYL)ETHER, Divalent metal cation transporter MntH, ... | | Authors: | Bozzi, A.T, Ray, S, Zimanyi, C.M, Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution structures with bound Mn 2+ and Cd 2+ map the metal import pathway in an Nramp transporter.
Elife, 12, 2023
|
|
6C4A
 
 | | Crystal structure of 3-nitropropionate modified isocitrate lyase from Mycobacterium tuberculosis with pyruvate | | Descriptor: | 3-NITROPROPANOIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kreitler, D.F, Ray, S, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Nitro Group as a Masked Electrophile in Covalent Enzyme Inhibition.
ACS Chem. Biol., 13, 2018
|
|
6C4C
 
 | | Crystal structure of 3-nitropropionate modified isocitrate lyase from Mycobacterium tuberculosis with glyoxylate and pyruvate | | Descriptor: | 3-NITROPROPANOIC ACID, GLYOXYLIC ACID, Isocitrate lyase 1, ... | | Authors: | Kreitler, D.F, Ray, S, Murkin, A.S, Gulick, A.M. | | Deposit date: | 2018-01-11 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Nitro Group as a Masked Electrophile in Covalent Enzyme Inhibition.
ACS Chem. Biol., 13, 2018
|
|
1CX8
 
 | | CRYSTAL STRUCTURE OF THE ECTODOMAIN OF HUMAN TRANSFERRIN RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SAMARIUM (III) ION, TRANSFERRIN RECEPTOR PROTEIN | | Authors: | Lawrence, C.M, Ray, S, Babyonyshev, M, Galluser, R, Borhani, D, Harrison, S.C. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the ectodomain of human transferrin receptor.
Science, 286, 1999
|
|
6QGK
 
 | | Structure of human Bcl-2 in complex with THIQ-phenyl pyrazole compound | | Descriptor: | 1-[2-[[(3~{S})-3-(aminomethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]phenyl]-~{N},~{N}-dibutyl-5-methyl-pyrazole-3-carboxamide, ACETATE ION, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGD
 
 | | Structure of human Mcl-1 in complex with thienopyrimidine inhibitor | | Descriptor: | 2-[(6-ethyl-5-phenyl-thieno[2,3-d]pyrimidin-4-yl)amino]-3-oxidanyl-propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGG
 
 | | Structure of human Bcl-2 in complex with analogue of ABT-737 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, [(3~{R})-3-[[4-[[4-[4-[[2-(4-chlorophenyl)phenyl]methyl]piperazin-1-yl]phenyl]carbonylsulfamoyl]-2-nitro-phenyl]amino]-4-phenylsulfanyl-butyl]-(2-hydroxy-2-oxoethyl)-dimethyl-azanium | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QFI
 
 | | Structure of human Mcl-1 in complex with BIM BH3 peptide | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QFQ
 
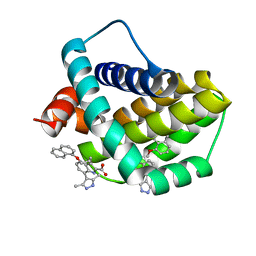 | | Structure of human Mcl-1 in complex with indole acid inhibitor | | Descriptor: | 7-(3,5-dimethyl-1~{H}-pyrazol-4-yl)-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QG8
 
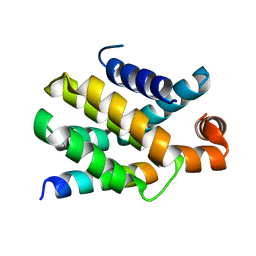 | | Structure of human Bcl-2 in complex with PUMA BH3 peptide | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, Bcl-2-binding component 3 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QFM
 
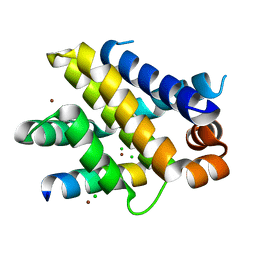 | | Structure of human Mcl-1 in complex with PUMA BH3 peptide | | Descriptor: | Bcl-2-binding component 3, CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGH
 
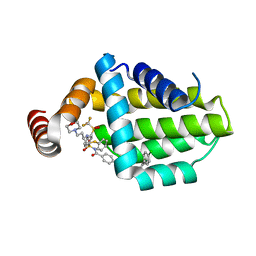 | | Structure of human Bcl-2 in complex with ABT-263 | | Descriptor: | 4-(4-{[2-(4-chlorophenyl)-5,5-dimethylcyclohex-1-en-1-yl]methyl}piperazin-1-yl)-N-[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
