5M63
 
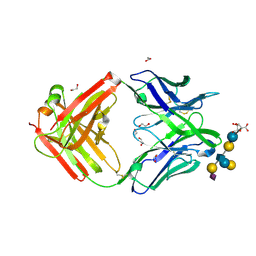 | | Crystal structure of group B Streptococcus type III DP2 oligosaccharide bound to Fab NVS-1-19-5 | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-[bis(oxidanyl)methyl]-5-(hydroxymethyl)oxolane-3,4-diol, 1,2-ETHANEDIOL, H chain of Fab NVS-1-19-5, ... | | Authors: | Carboni, F, Adamo, R, Veggi, D, Rappuoli, R, Malito, E, Margarit, I.R, Berti, F. | | Deposit date: | 2016-10-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of a protective epitope of group B Streptococcus type III capsular polysaccharide.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1YS5
 
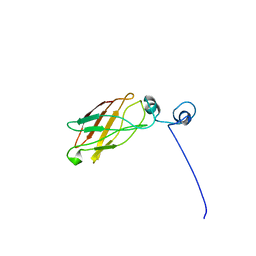 | | Solution structure of the antigenic domain of GNA1870 of Neisseria meningitidis | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Savino, S, Masignani, V, Pizza, M, Scarselli, M, Swennen, E, Romagnoli, G, Veggi, D, Banci, L, Rappuoli, R. | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis
J.Biol.Chem., 281, 2006
|
|
1JI4
 
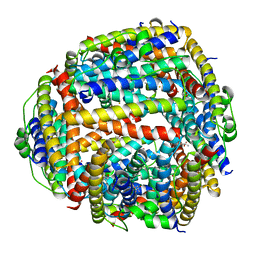 | | NAP protein from helicobacter pylori | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE (III) ION, NEUTROPHIL-ACTIVATING PROTEIN A, ... | | Authors: | Zanotti, G, Papinutto, E, Dundon, W.G, Battistutta, R, Seveso, M, Del Giudice, G, Rappuoli, R, Montecucco, C. | | Deposit date: | 2001-06-29 | | Release date: | 2002-10-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Neutrophil-activating Protein from Helicobacter pylori
J.Mol.Biol., 323, 2002
|
|
2KC0
 
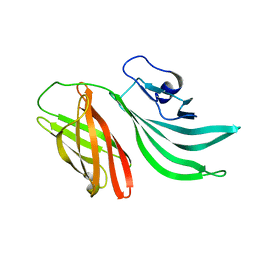 | | Solution structure of the factor H binding protein | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Veggi, D, Dragonetti, S, Savino, S, Scarselli, M, Romagnoli, G, Pizza, M, Banci, L, Rappuoli, R. | | Deposit date: | 2008-12-13 | | Release date: | 2009-02-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Factor H-binding Protein, a Survival Factor and Protective Antigen of Neisseria meningitidis
J.Biol.Chem., 284, 2009
|
|
3RKI
 
 | | Structural basis for immunization with post-fusion RSV F to elicit high neutralizing antibody titers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F0 | | Authors: | Swanson, K.A, Settembre, E.C, Shaw, C.A, Dey, A.K, Rappuoli, R, Mandl, C.W, Dormitzer, P.D, Carfi, A. | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4E9L
 
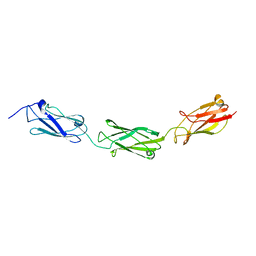 | | FdeC, a Novel Broadly Conserved Escherichia coli Adhesin Eliciting Protection against Urinary Tract Infections | | Descriptor: | Attaching and effacing protein, pathogenesis factor | | Authors: | Spraggon, G, Nesta, B, Alteri, C, Gomes Moriel, D, Rosini, R, Veggi, D, Smith, S, Bertoldi, I, Pastorello, I, Ferlenghi, I, Fontana, M.R, Frankel, G, Mobley, H.L.T, Rappuoli, R, Pizza, M, Serino, L, Soriana, M. | | Deposit date: | 2012-03-21 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections.
MBio, 3, 2012
|
|
2X9Z
 
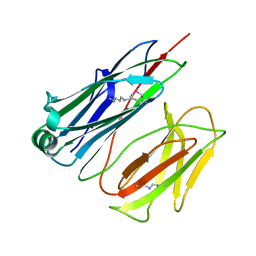 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9W
 
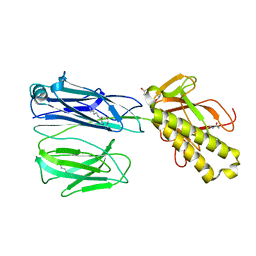 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, GLYCEROL | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9X
 
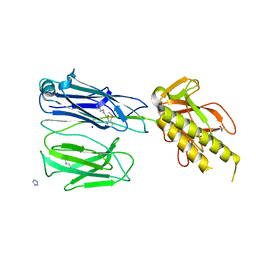 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, IMIDAZOLE, SODIUM ION | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
2X9Y
 
 | | STRUCTURE OF THE PILUS BACKBONE (RRGB) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN | | Authors: | Spraggon, G, Koesema, E, Scarselli, M, Malito, E, Biagini, M, Norais, N, Emolo, C, Barocchi, M.A, Giusti, F, Hilleringmann, M, Rappuoli, R, Lesley, S, Covacci, A, Masignani, V, Ferlenghi, I. | | Deposit date: | 2010-03-25 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Supramolecular Organization of the Repetitive Backbone Unit of the Streptococcus Pneumoniae Pilus.
Plos One, 5, 2010
|
|
1HTL
 
 | |
6Y54
 
 | | Crystal structure of a Neisseria meningitidis serogroup A capsular oligosaccharide bound to a functional Fab | | Descriptor: | 1,2-ETHANEDIOL, Fab A1.1 H chain, Fab A1.1 L chain, ... | | Authors: | Dello Iacono, L, Henriques, P, Adamo, R. | | Deposit date: | 2020-02-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structure of a protective epitope reveals the importance of acetylation of Neisseria meningitidis serogroup A capsular polysaccharide.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XZW
 
 | |
1LTG
 
 | |
1LT4
 
 | | HEAT-LABILE ENTEROTOXIN MUTANT S63K | | Descriptor: | HEAT-LABILE ENTEROTOXIN, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Van Den Akker, F, Hol, W.G.J. | | Deposit date: | 1997-04-14 | | Release date: | 1997-06-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a non-toxic mutant of heat-labile enterotoxin, which is a potent mucosal adjuvant.
Protein Sci., 6, 1997
|
|
2FJC
 
 | | Crystal structure of antigen TpF1 from Treponema pallidum | | Descriptor: | Antigen TpF1, FE (III) ION | | Authors: | Thumiger, A, Polenghi, A, Papinutto, E, Battistutta, R, Montecucco, C, Zanotti, G. | | Deposit date: | 2006-01-02 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of antigen TpF1 from Treponema pallidum.
Proteins, 62, 2006
|
|
7S6L
 
 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (conformation 3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7S6K
 
 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7S6J
 
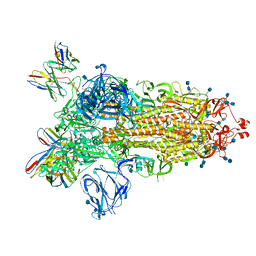 | | J08 fragment antigen binding in complex with SARS-CoV-2-6P-Mut2 S protein (conformation 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, J08 fragment antigen binding heavy chain variable domain, ... | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SBU
 
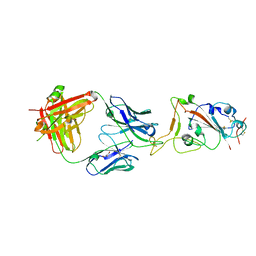 | |
7S6I
 
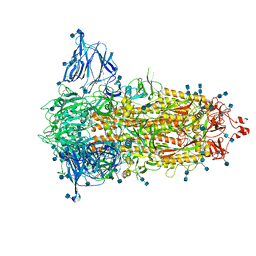 | | SARS-CoV-2-6P-Mut2 S protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Torres, J.L, Ward, A.B. | | Deposit date: | 2021-09-14 | | Release date: | 2022-05-11 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1O9R
 
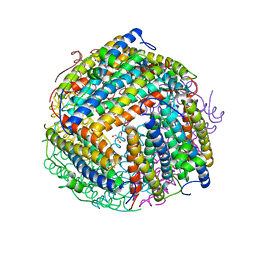 | | The X-ray crystal structure of Agrobacterium tumefaciens Dps, a member of the family that protect DNA without binding | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AGROBACTERIUM TUMEFACIENS DPS, ... | | Authors: | Ilari, A, Ceci, P, Chiancone, E. | | Deposit date: | 2002-12-18 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Dps Protein of Agrobacterium Tumefaciens Does not Bind to DNA But Protects It Toward Oxidative Cleavage: X-Ray Crystal Structure, Iron Binding, and Hydroxyl-Radical Scavenging Properties
J.Biol.Chem., 278, 2003
|
|
4CJD
 
 | | Crystal structure of Neisseria meningitidis trimeric autotransporter and vaccine antigen NadA | | Descriptor: | IODIDE ION, NADA | | Authors: | Malito, E, Biancucci, M, Spraggon, G, Bottomley, M.J. | | Deposit date: | 2013-12-19 | | Release date: | 2014-11-26 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.056 Å) | | Cite: | Structure of the Meningococcal Vaccine Antigen Nada and Epitope Mapping of a Bactericidal Antibody.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2XTL
 
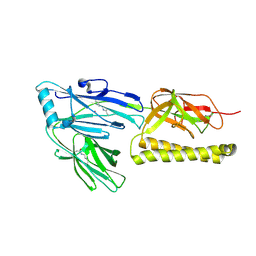 | | Structure of the major pilus backbone protein from Streptococcus Agalactiae | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, POTASSIUM ION | | Authors: | Rinauda, D, Gourlay, L.J, Soriano, M, Grandi, G, Bolognesi, M. | | Deposit date: | 2010-10-11 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Approach to Rationally Design a Chimeric Protein for an Effective Vaccine Against Group B Streptococcus Infections.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
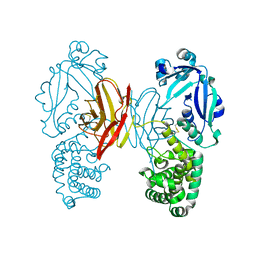
4AE0
 
 | |
