2VHE
 
 | | PglD-CoA complex: An acetyl transferase from Campylobacter jejuni | | Descriptor: | ACETYLTRANSFERASE, COENZYME A, SULFATE ION | | Authors: | Rangarajan, E.S, Ruane, K.M, Sulea, T, Watson, D.C, Proteau, A, Leclerc, S, Cygler, M, Matte, A, Young, N.M. | | Deposit date: | 2007-11-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Active Site Residues of Pgld, an N-Acetyltransferase from the Bacillosamine Synthetic Pathway Required for N-Glycan Synthesis in Campylobacter Jejuni
Biochemistry, 47, 2008
|
|
1LKZ
 
 | | Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli. | | Descriptor: | Ribose 5-phosphate isomerase A | | Authors: | Rangarajan, E.S, Sivaraman, J, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-04-26 | | Release date: | 2002-05-08 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of D-ribose-5-phosphate isomerase (RpiA) from Escherichia coli
Proteins, 48, 2002
|
|
3BFP
 
 | | Crystal Structure of apo-PglD from Campylobacter jejuni | | Descriptor: | Acetyltransferase, CITRATE ANION | | Authors: | Rangarajan, E.S, Watson, D.C, Leclerc, S, Proteau, A, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-11-22 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Active Site Residues of PglD, an N-Acetyltransferase from the Bacillosamine Synthetic Pathway Required for N-Glycan Synthesis in Campylobacter jejuni.
Biochemistry, 47, 2008
|
|
2AHW
 
 | | Crystal Structure of Acyl-CoA transferase from E. coli O157:H7 (YdiF)-thioester complex with CoA- 2 | | Descriptor: | COENZYME A, putative enzyme YdiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cylger, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
2AHV
 
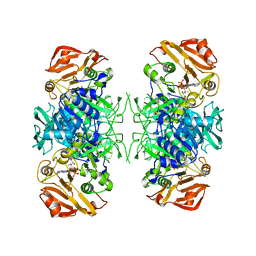 | | Crystal Structure of Acyl-CoA transferase from E. coli O157:H7 (YdiF)-thioester complex with CoA- 1 | | Descriptor: | COENZYME A, putative enzyme YdiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
2AHU
 
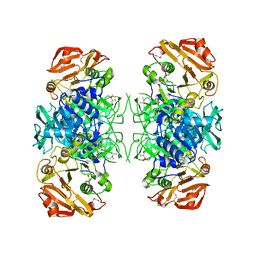 | | Crystal structure of Acyl-CoA transferase (YdiF) apoenzyme from Escherichia coli O157:H7. | | Descriptor: | putative enzyme ydiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
3HBM
 
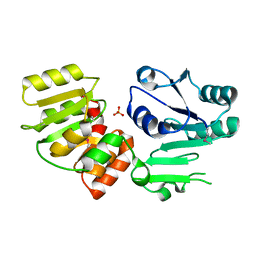 | | Crystal Structure of PseG from Campylobacter jejuni | | Descriptor: | SULFATE ION, UDP-sugar hydrolase | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
3HBN
 
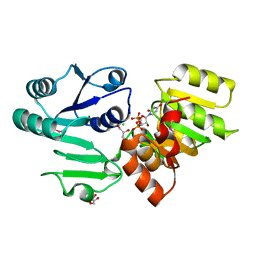 | | Crystal structure PseG-UDP complex from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, GLYCEROL, UDP-sugar hydrolase, ... | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
7UUW
 
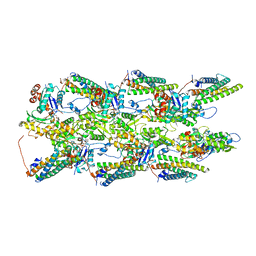 | | Cryogenic electron microscopy 3D map of F-actin bound by the Actin Binding Domain of alpha-catenin ortholog, HMP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The nematode alpha-catenin ortholog, HMP1, has an extended alpha-helix when bound to actin filaments.
J.Biol.Chem., 299, 2022
|
|
7UTJ
 
 | | Cryogenic electron microscopy 3D map of F-actin bound by human dimeric alpha-catenin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
7UXF
 
 | | Cryogenic electron microscopy 3D map of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
3LGE
 
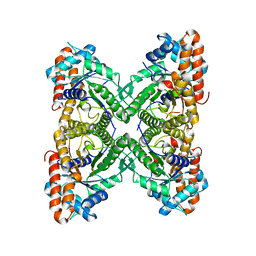 | | Crystal structure of rabbit muscle aldolase-SNX9 LC4 complex | | Descriptor: | Fructose-bisphosphate aldolase A, Sorting nexin-9 | | Authors: | Rangarajan, E.S, Park, H, Fortin, E, Sygusch, J, Izard, T. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
J.Biol.Chem., 285, 2010
|
|
3SMZ
 
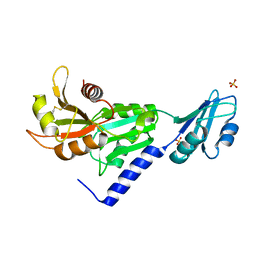 | |
1SBZ
 
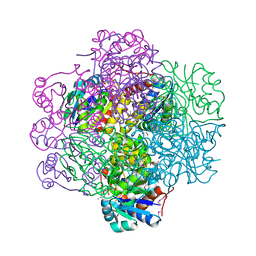 | | Crystal Structure of dodecameric FMN-dependent Ubix-like Decarboxylase from Escherichia coli O157:H7 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Probable aromatic acid decarboxylase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Tocilj, A, Hung, L.-W, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-02-11 | | Release date: | 2004-10-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a dodecameric FMN-dependent UbiX-like decarboxylase (Pad1) from Escherichia coli O157: H7.
Protein Sci., 13, 2004
|
|
1XK7
 
 | | Crystal Structure- C2 form- of Escherichia coli Crotonobetainyl-CoA: carnitine CoA transferase (CaiB) | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1XVT
 
 | | Crystal Structure of Native CaiB in complex with coenzyme A | | Descriptor: | COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-10-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1XK6
 
 | | Crystal Structure- P1 form- of Escherichia coli Crotonobetainyl-CoA: carnitine CoA Transferase (CaiB) | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1XVU
 
 | | Crystal Structure of CaiB mutant D169A in complex with Coenzyme A | | Descriptor: | COENZYME A, Crotonobetainyl-CoA:carnitine CoA-transferase | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-10-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
1XVV
 
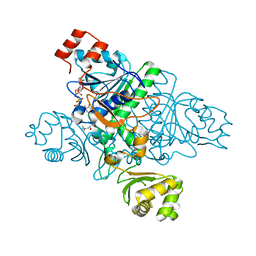 | | Crystal Structure of CaiB mutant D169A in complex with carnitinyl-CoA | | Descriptor: | Crotonobetainyl-CoA:carnitine CoA-transferase, L-CARNITINYL-COA INNER SALT | | Authors: | Rangarajan, E.S, Li, Y, Iannuzzi, P, Cygler, M, Matte, A. | | Deposit date: | 2004-10-28 | | Release date: | 2005-03-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Escherichia coli Crotonobetainyl-CoA: Carnitine CoA-Transferase (CaiB) and Its Complexes with CoA and Carnitinyl-CoA.
Biochemistry, 44, 2005
|
|
2FLO
 
 | |
2FPU
 
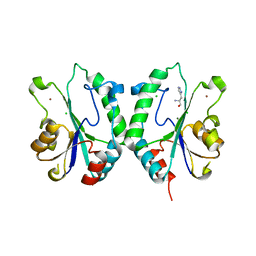 | | Crystal Structure of the N-terminal domain of E.coli HisB- Complex with histidinol | | Descriptor: | CHLORIDE ION, Histidine biosynthesis bifunctional protein hisB, L-histidinol, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
2FPX
 
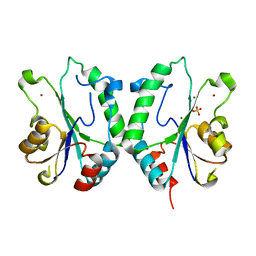 | | Crystal Structure of the N-terminal Domain of E.coli HisB- Sulfate complex. | | Descriptor: | Histidine biosynthesis bifunctional protein hisB, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
2FPW
 
 | | Crystal Structure of the N-terminal Domain of E.coli HisB- Phosphoaspartate intermediate. | | Descriptor: | CALCIUM ION, Histidine biosynthesis bifunctional protein hisB, ZINC ION | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
2FPS
 
 | | Crystal structure of the N-terminal domain of E.coli HisB- Apo Ca model. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Histidine biosynthesis bifunctional protein hisB, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
2FPR
 
 | | Crystal structure the N-terminal domain of E. coli HisB. Apo Mg model. | | Descriptor: | BROMIDE ION, Histidine biosynthesis bifunctional protein hisB, MAGNESIUM ION, ... | | Authors: | Rangarajan, E.S, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2006-01-17 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway.
J.Biol.Chem., 281, 2006
|
|
