3GT2
 
 | | Crystal Structure of the P60 Domain from M. avium paratuberculosis Antigen MAP1272c | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein, SULFATE ION | | Authors: | Ramyar, K.X, Lingle, C.K, McWhorter, W.J, Bouyain, S, Bannantine, J.P, Geisbrecht, B.V. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Two P60-Family Antigens from Mycobacterium Avium Paratuberculosis
To be Published
|
|
3I86
 
 | | Crystal structure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 | | Descriptor: | ISOPROPYL ALCOHOL, Putative uncharacterized protein | | Authors: | Ramyar, K.X, Lingle, C.K, McWhorter, W.J, Bouyain, S, Bannantine, J.P, Geisbrecht, B.V. | | Deposit date: | 2009-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atypical structural features of two P60 family members from Mycobacterium avium subspecies paratuberculosis
To be Published
|
|
5JXU
 
 | | Structural basis for the catalytic activity of Thermomonospora curvata heme-containing DyP-type peroxidase. | | Descriptor: | Dyp-type peroxidase family, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ramyar, K.X, Carlson, E.A, Li, P, Geisbrecht, B.V. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Identification of Surface-Exposed Protein Radicals and A Substrate Oxidation Site in A-Class Dye-Decolorizing Peroxidase from Thermomonospora curvata.
ACS Catal, 6, 2016
|
|
6AZP
 
 | |
4NZL
 
 | | Extracellular proteins of Staphylococcus aureus inhibit the neutrophil serine proteases | | Descriptor: | Neutrophil elastase, Uncharacterized protein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Stapels, D.A.C, von Koeckritz-Blickwede, M, Ramyar, K.X, Bischoff, M, Milder, F, Ruyken, M, Scheepmaker, L, McWhorter, W.J, Herrmann, M, van Kessel, K.P.M, Geisbrecht, B.V, Rooijakkers, S.H.M. | | Deposit date: | 2013-12-12 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Staphylococcus aureus secretes a unique class of neutrophil serine protease inhibitors.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1N8Z
 
 | | Crystal structure of extracellular domain of human HER2 complexed with Herceptin Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Herceptin Fab (antibody) - heavy chain, Herceptin Fab (antibody) - light chain, ... | | Authors: | Cho, H.-S, Mason, K, Ramyar, K.X, Stanley, A.M, Gabelli, S.B, Denney Jr, D.W, Leahy, D.J. | | Deposit date: | 2002-11-21 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Extracellular Region of HER2 Alone and in Complex with the Herceptin Fab
Nature, 421, 2003
|
|
1N8Y
 
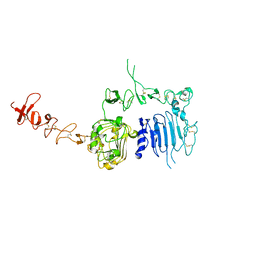 | | Crystal structure of the extracellular region of rat HER2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, protooncoprotein | | Authors: | Cho, H.-S, Mason, K, Ramyar, K.X, Stanley, A.M, Gabelli, S.B, Denney Jr, D.W, Leahy, D.J. | | Deposit date: | 2002-11-21 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Extracellular Region of HER2 Alone and in complex with the Herceptin Fab
Nature, 421, 2003
|
|
3V4X
 
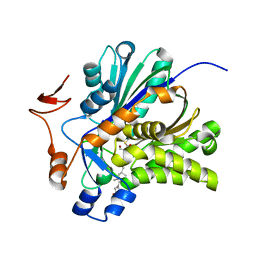 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthase, mvaS, by Hymeglusin | | Descriptor: | (7R,12R,13R)-13-formyl-12,14-dihydroxy-3,5,7-trimethyltetradeca-2,4-dienoic acid, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
3V4N
 
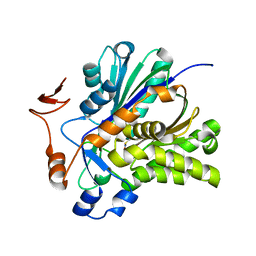 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthatse, mvaS, by Hymeglusin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
6D69
 
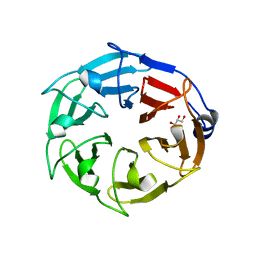 | |
7TE3
 
 | | Crystal Structure of a Double Loop Deletion Mutant in gC1qR/C1qBP/HABP-1 | | Descriptor: | Complement component 1 Q subcomponent-binding protein, mitochondrial | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2022-01-04 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | gC1qR/C1qBP/HABP-1: Structural Analysis of the Trimeric Core Region, Interactions With a Novel Panel of Monoclonal Antibodies, and Their Influence on Binding to FXII.
Front Immunol, 13, 2022
|
|
2NOJ
 
 | | Crystal structure of Ehp / C3d complex | | Descriptor: | Complement C3, Efb homologous protein | | Authors: | Hammel, M, Geisbrecht, B.V. | | Deposit date: | 2006-10-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Characterization of Ehp, a Secreted Complement Inhibitory Protein from Staphylococcus aureus.
J.Biol.Chem., 282, 2007
|
|
5UZU
 
 | | Immune evasion by a Staphylococcal Peroxidase Inhibitor that blocks myeloperoxidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | de Jong, N, Geisbrecht, B.V, van Strijp, J, Haas, P, Nijland, R, Ramyar, K, Fevre, C, Guerra, F, Voyich-Kane, J, van Kessel, C, Garcia, B. | | Deposit date: | 2017-02-27 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Immune evasion by a staphylococcal inhibitor of myeloperoxidase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4H6H
 
 | |
4H6I
 
 | |
3NMS
 
 | |
3OHX
 
 | |
5WKQ
 
 | | 2.10 A resolution structure of IpaB (residues 74-242) from Shigella flexneri | | Descriptor: | Invasin IpaB | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Picking, W.L, Picking, W.D. | | Deposit date: | 2017-07-25 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Using disruptive insertional mutagenesis to identify the in situ structure-function landscape of the Shigella translocator protein IpaB.
Protein Sci., 27, 2018
|
|
3L3O
 
 | |
3L5N
 
 | |
1S78
 
 | | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, ... | | Authors: | Franklin, M.C, Carey, K.D, Vajdos, F.F, Leahy, D.J, de Vos, A.M, Sliwkowski, M.X. | | Deposit date: | 2004-01-29 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex.
Cancer Cell, 5, 2004
|
|
3T46
 
 | |
3T49
 
 | |
3T47
 
 | |
3T48
 
 | |
