5OYN
 
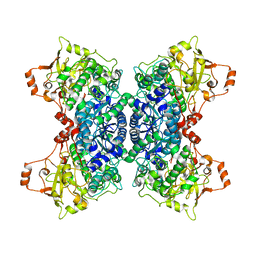 | | Crystal structure of D-xylonate dehydratase in holo-form | | Descriptor: | Dehydratase, IlvD/Edd family, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rahman, M.M, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2017-09-11 | | Release date: | 2018-01-24 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of D-xylonate dehydratase reveals functional features of enzymes from the Ilv/ED dehydratase family.
Sci Rep, 8, 2018
|
|
6OFQ
 
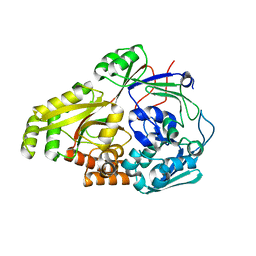 | | ABC transporter-associated periplasmic binding protein DppA from Helicobacter pylori in complex with peptide STSA | | Descriptor: | Heme-binding protein A / AI-2 binding protein A, SER-THR-SER-ALA | | Authors: | Rahman, M.M, Machuca, M.A, Khan, M.F, Barlow, C.K, Schittenhelm, R.B, Roujeinikova, A. | | Deposit date: | 2019-03-31 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
J.Bacteriol., 201, 2019
|
|
6PU3
 
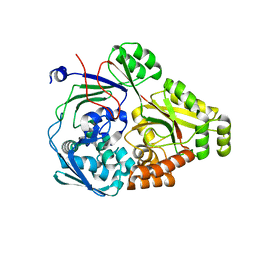 | | ABC transporter-associated periplasmic binding protein DppA from Helicobacter pylori | | Descriptor: | Heme-binding protein A, SER-THR-SER-ALA | | Authors: | Rahman, M.M, Machuca, M.A, Khan, M.F, Barlow, C.K, Schittenhelm, R.B, Roujeinikova, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Unexpected Specificity of ABC Transporter-Associated Substrate-Binding Protein DppA from Helicobacter pylori.
J.Bacteriol., 201, 2019
|
|
5J83
 
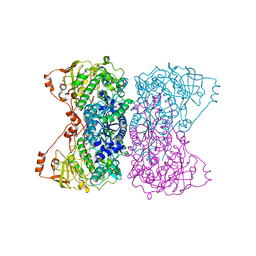 | |
5J85
 
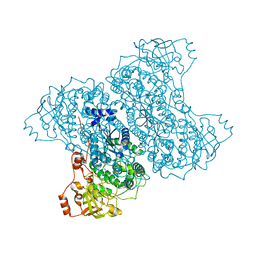 | | Ser480Ala mutant of L-arabinonate dehydratase | | Descriptor: | Dihydroxyacid dehydratase/phosphogluconate dehydratase, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION | | Authors: | Rahman, M.M, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2016-04-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of a Bacterial l-Arabinonate Dehydratase Contains a [2Fe-2S] Cluster.
ACS Chem. Biol., 12, 2017
|
|
5J84
 
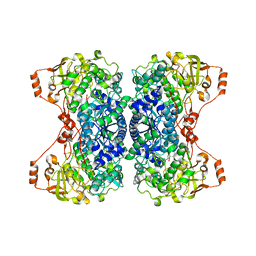 | |
6UWZ
 
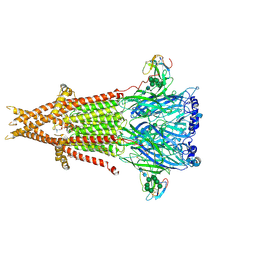 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with alpha-bungarotoxin | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Rahman, M.M, Teng, J, Worrell, B.T, Noveillo, C.M, Lee, M, Karlin, A, Stowell, M, Hibbs, R.E. | | Deposit date: | 2019-11-06 | | Release date: | 2020-04-15 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the Native Muscle-type Nicotinic Receptor and Inhibition by Snake Venom Toxins.
Neuron, 106, 2020
|
|
7SMR
 
 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with carbachol, desensitized state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SMQ
 
 | | Cryo-EM structure of Torpedo acetylcholine receptor in apo form with added cholesterol | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SMM
 
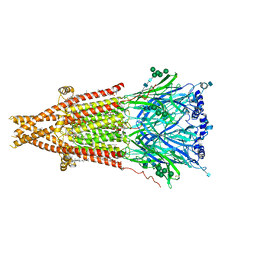 | | Cryo-EM structure of Torpedo acetylcholine receptor in apo form | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SMT
 
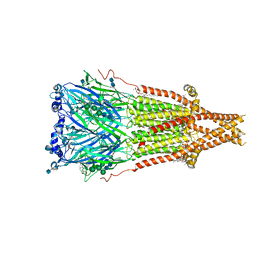 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with d-tubocurarine and carbachol | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SMS
 
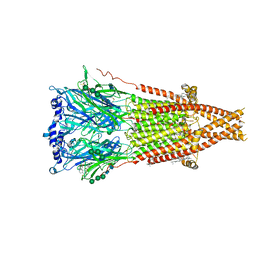 | | Cryo-EM structure of Torpedo acetylcholine receptor in complex with d-tubocurarine | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine receptor subunit alpha, ... | | Authors: | Rahman, M.M, Basta, T, Teng, J, Lee, M, Worrell, B.T, Stowell, M.H.B, Hibbs, R.E. | | Deposit date: | 2021-10-26 | | Release date: | 2022-03-09 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural mechanism of muscle nicotinic receptor desensitization and block by curare.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5GJ3
 
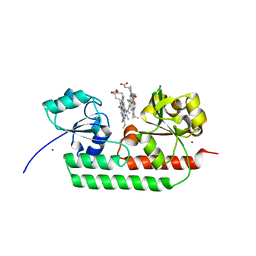 | | Periplasmic heme-binding protein RhuT from Roseiflexus sp. RS-1 in two-heme bound form (holo-2) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Periplasmic binding protein, ZINC ION | | Authors: | Rahman, M.M, Naoe, Y, Nakamura, N, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems.
Proteins, 85, 2017
|
|
7W7C
 
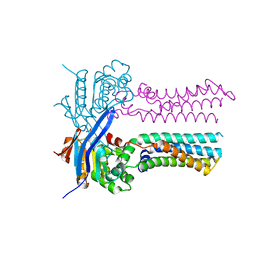 | | Heme exporter in the unliganded form | | Descriptor: | Putative ABC transport system integral membrane protein, Putative ABC transport system, ATP-binding protein, ... | | Authors: | Rahman, M.M, Hisano, T, Nakamura, H, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5Y8B
 
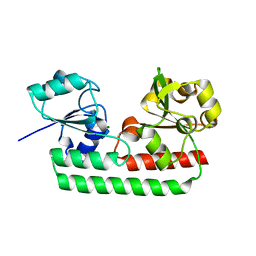 | | Periplasmic heme-binding protein RhuT from Roseiflexus sp. RS-1 in apo form | | Descriptor: | MAGNESIUM ION, Periplasmic binding protein | | Authors: | Rahman, M.M, Naoe, Y, Nakamura, N, Doi, A, Shiro, Y, Sugimoto, H. | | Deposit date: | 2017-08-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems
Proteins, 85, 2017
|
|
5GIZ
 
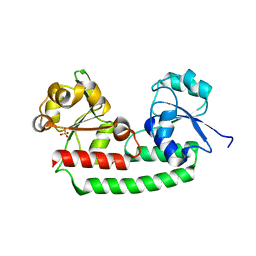 | | Periplasmic heme-binding protein BhuT in apo form | | Descriptor: | CHLORIDE ION, Putative hemin transport system, substrate-binding protein, ... | | Authors: | Nakamura, N, Naoe, Y, Rahman, M.M, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems.
Proteins, 85, 2017
|
|
5LN8
 
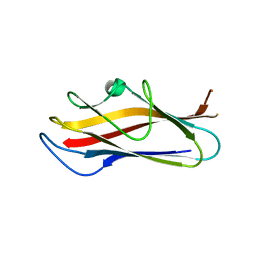 | | Crystal structure of self-complemented MyfA, the major subunit of Myf fimbriae from Yersinia enterocolitica, in complex with galactose | | Descriptor: | Fimbrial protein MyfA,Fimbrial protein MyfA, beta-D-galactopyranose | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
5LN4
 
 | | Crystal structure of self-complemented PsaA, the major subunit of pH 6 antigen from Yersinia pests, in complex with choline | | Descriptor: | CHOLINE ION, pH 6 antigen,pH 6 antigen | | Authors: | Pakharukova, N.A, Roy, S, Rahman, M.M, Tuitilla, M, Zavialov, A.V. | | Deposit date: | 2016-08-03 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for Myf and Psa fimbriae-mediated tropism of pathogenic strains of Yersinia for host tissues.
Mol.Microbiol., 102, 2016
|
|
7W78
 
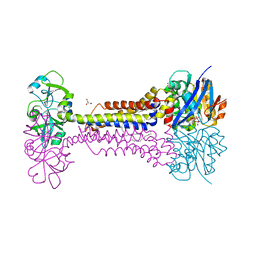 | | Heme exporter HrtBA in complex with Mg-AMPPNP | | Descriptor: | ACETATE ION, DODECANE, GLYCEROL, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W79
 
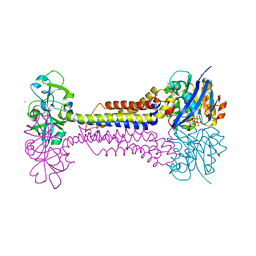 | | Heme exporter HrtBA in complex with Mn-AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, Putative ABC transport system integral membrane protein, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7B
 
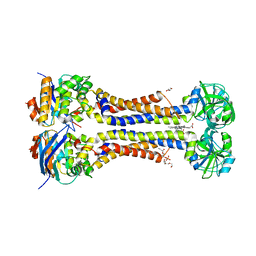 | | Heme exporter HrtBA in complex with protoporphyrin IX containing manganese(III), high resolution data | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING MN, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7A
 
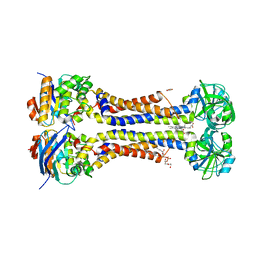 | | Heme exporter in complex with Mn-containing protoporphyrin IX, Mn-anomalous data | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7D
 
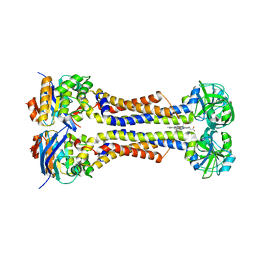 | | Heme exporter HrtBA in complex with heme | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative ABC transport system integral membrane protein, Putative ABC transport system, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shiro, Y, Shirouzu, M. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5Y89
 
 | | Periplasmic heme-binding protein BhuT in complex with one heme (holo-1) | | Descriptor: | ACETATE ION, PROTOPORPHYRIN IX CONTAINING FE, Putative hemin transport system, ... | | Authors: | Naoe, Y, Nakamura, N, Rahman, M.M, Shiro, Y, Sugimoto, H. | | Deposit date: | 2017-08-20 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems
Proteins, 85, 2017
|
|
5Y8A
 
 | | Periplasmic heme-binding protein BhuT in complex with two hemes (holo-2 form) | | Descriptor: | CALCIUM ION, PROTOPORPHYRIN IX CONTAINING FE, Putative hemin transport system, ... | | Authors: | Nakamura, N, Naoe, Y, Rahman, M.M, Shiro, Y, Sugimoto, H. | | Deposit date: | 2017-08-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems
Proteins, 85, 2017
|
|
