1PU6
 
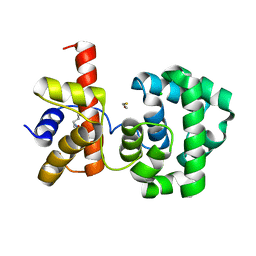 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-METHYLADENINE DNA GLYCOSYLASE, BETA-MERCAPTOETHANOL, ... | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PU7
 
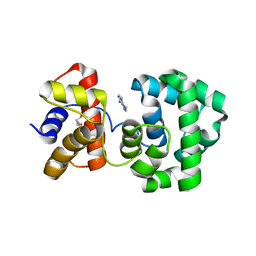 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 3,9-dimethyladenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 6-AMINO-3,9-DIMETHYL-9H-PURIN-3-IUM, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
1PU8
 
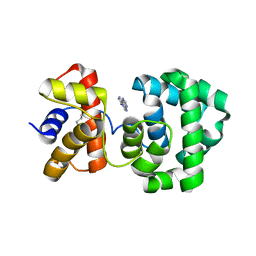 | | Crystal structure of H.pylori 3-methyladenine DNA glycosylase (MagIII) bound to 1,N6-ethenoadenine | | Descriptor: | 3-METHYLADENINE DNA GLYCOSYLASE, 3H-IMIDAZO[2,1-I]PURINE, BETA-MERCAPTOETHANOL | | Authors: | Eichman, B.F, O'Rourke, E.J, Radicella, J.P, Ellenberger, T. | | Deposit date: | 2003-06-24 | | Release date: | 2003-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases
Embo J., 22, 2003
|
|
6GW7
 
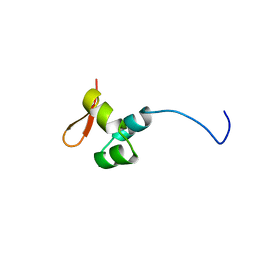 | | The CTD of HpDprA, a DNA binding Winged Helix domain which do not bind dsDNA | | Descriptor: | DNA protecting protein DprA | | Authors: | Lisboa, J, Celma, L, Sanchez, D, Marquis, M, Andreani, J, Guerois, R, Ochsenbein, F, Durand, D, Marsin, S, Cuniasse, P, Radicella, J.P, Quevillon-Cheruel, S. | | Deposit date: | 2018-06-22 | | Release date: | 2019-04-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of HpDprA is a DNA-binding winged helix domain that does not bind double-stranded DNA.
Febs J., 286, 2019
|
|
7P0H
 
 | | Crystal structure of Helicobacter pylori ComF fused to an artificial alphaREP crystallization helper(named B2) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, GLYCEROL, Helicobacter pylori ComF fused to an artificial alphaREP crystallization helper (named B2), ... | | Authors: | Celma, L, Walbott, H, Legrand, P, Quevillon-Cheruel, S. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2022-04-27 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | ComFC mediates transport and handling of single-stranded DNA during natural transformation.
Nat Commun, 13, 2022
|
|
5MLL
 
 | |
6EHI
 
 | | NucT from Helicobacter pylori | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Celma, L, Li de la Sierra-Gallay, I, Quevillon-Cheruel, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis for the substrate selectivity of Helicobacter pylori NucT nuclease activity.
PLoS ONE, 12, 2017
|
|
