7QPB
 
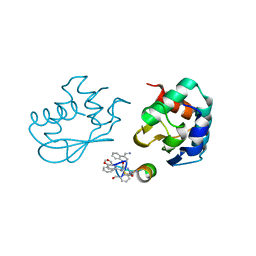 | | Catalytic C-lobe of the HECT-type ubiquitin ligase E6AP in complex with a hybrid foldamer-peptide macrocycle | | Descriptor: | Isoform I of Ubiquitin-protein ligase E3A, hybrid foldamer-peptide macrocycle | | Authors: | Dengler, S, Howard, R.T, Morozov, V, Tsiamantas, C, Douat, C, Suga, H, Huc, I. | | Deposit date: | 2022-01-03 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Display Selection of a Hybrid Foldamer-Peptide Macrocycle.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7JR8
 
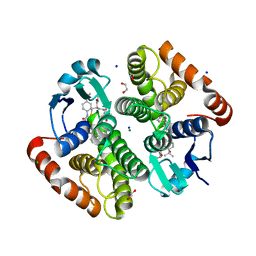 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7JR6
 
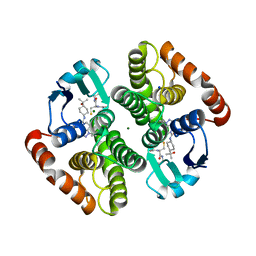 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1-(3-fluorophenyl)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
4W4S
 
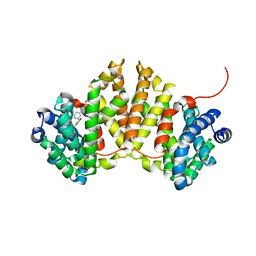 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum in complex with BPH-629 | | Descriptor: | Uncharacterized protein blr2150, [2-(3-DIBENZOFURAN-4-YL-PHENYL)-1-HYDROXY-1-PHOSPHONO-ETHYL]-PHOSPHONIC ACID | | Authors: | Liu, W, Zheng, Y, Huang, C.H, Guo, R.T. | | Deposit date: | 2014-08-15 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4W4R
 
 | | Crystal structure of ent-kaurene synthase BJKS from bradyrhizobium japonicum | | Descriptor: | Uncharacterized protein blr2150 | | Authors: | Liu, W, Zheng, Y, Huang, C.H, Ko, T.P, Guo, R.T. | | Deposit date: | 2014-08-15 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4WOQ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric | | Descriptor: | 2-KETOBUTYRIC ACID, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-16 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with ketobutyric
to be published
|
|
4WOZ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
to be published
|
|
4WNQ
 
 | | THE MOLECULAR BASES OF DELTA/ALPHA-BETA T-CELL MEDIATED ANTIGEN RECOGNITION | | Descriptor: | TCR Variable Beta 2 (TRBV20) chain and TCR constant Beta chain, TCR Variable Delta 1 chain and TCR constant Alpha chain | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-14 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
4WO4
 
 | | The molecular bases of Delta/Alpha beta T cell-mediated antigen recognition. | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pellicci, D.G, Uldrich, A.P, Le Nours, J, Ross, F, Chabrol, E, Eckle, S.B.G, de Boer, R, Lim, R.T, McPherson, K, Besra, G, Howell, A.R, Moretta, L, McCluskey, J, Heemskerk, M.H.M, Gras, S, Rossjohn, J, Godfrey, D.I. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular bases of delta / alpha beta T cell-mediated antigen recognition.
J.Exp.Med., 211, 2014
|
|
4WW2
 
 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8, Beta Chain-TRBV7-8, Antigen-presenting glycoprotein CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, S, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-10 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4WW1
 
 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8 and Beta Chain-TRBV7-8 | | Descriptor: | TCR Alpha Chain-TRAV21-TRAJ8, TCR Beta Chain-TRBV7-8 | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Lim, R.T, Besra, G, Howell, A.R, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-10 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
6DKQ
 
 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 | | Descriptor: | Heme-binding protein Shr, SULFATE ION | | Authors: | Macdonald, R, Cascio, D, Collazo, M.J, Clubb, R.T. | | Deposit date: | 2018-05-30 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Streptococcus pyogenes Shr protein captures human hemoglobin using two structurally unique binding domains.
J.Biol.Chem., 293, 2018
|
|
6DN1
 
 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1151 SPLIT RNA | | Descriptor: | 10-(6-carboxyhexyl)-8-(cyclopentylamino)-2,4-dihydroxy-7-methylbenzo[g]pteridin-10-ium, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
6GSQ
 
 | | Oxidised copper nitrite reductase from Achromobacter cycloclastes determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION | | Authors: | Halsted, T.P, Yamashita, K, Gopalasingam, C.C, Shenoy, R.T, Hirata, K, Ago, H, Ueno, G, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GT2
 
 | | Reduced copper nitrite reductase from Achromobacter cycloclastes determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION | | Authors: | Halsted, T.P, Yamashita, K, Gopalasingam, C.C, Shenoy, R.T, Hirata, K, Ago, H, Ueno, G, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
6GT0
 
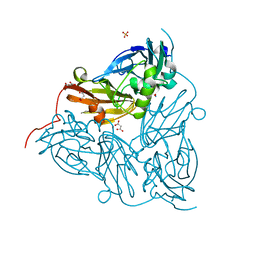 | | Nitrite-bound copper nitrite reductase from Achromobacter cycloclastes determined by serial femtosecond rotation crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, MALONATE ION, ... | | Authors: | Halsted, T.P, Yamashita, K, Gopalasingam, C.C, Shenoy, R.T, Hirata, K, Ago, H, Ueno, G, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2018-06-15 | | Release date: | 2019-07-03 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Catalytically important damage-free structures of a copper nitrite reductase obtained by femtosecond X-ray laser and room-temperature neutron crystallography.
Iucrj, 6, 2019
|
|
4Z2A
 
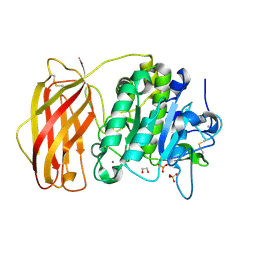 | | Crystal structure of unglycosylated apo human furin @1.89A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Furin, ... | | Authors: | Gampe, R.T, Pearce, K, Reid, R. | | Deposit date: | 2015-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | BacMam production and crystal structure of nonglycosylated apo human furin at 1.89A resolution
Acta Crystallogr.,Sect.F, 2019
|
|
6YSE
 
 | |
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7M1L
 
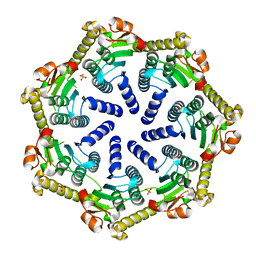 | | Crystal structure of Pseudomonas aeruginosa ClpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, PHOSPHATE ION | | Authors: | Hall, B.M, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
7B1O
 
 | | Crystal structure of the indoleamine 2,3-dioxygenase 1 (IDO1) in complex with compound 22 | | Descriptor: | 4-chloranyl-N-[(1R)-1-[(1S,5R)-3-quinolin-4-yloxy-6-bicyclo[3.1.0]hexanyl]propyl]benzamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lammens, A, Krapp, S, Lewis, R.T, Hamilton, M.M. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme.
J.Med.Chem., 64, 2021
|
|
7MPK
 
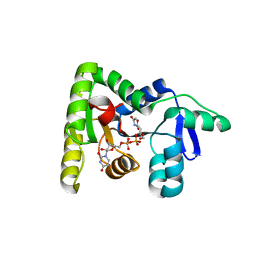 | | Crystal structure of TagA with UDP-GlcNAc | | Descriptor: | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Martinez, O.E, Cascio, D, Clubb, R.T. | | Deposit date: | 2021-05-04 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Insight into the molecular basis of substrate recognition by the wall teichoic acid glycosyltransferase TagA.
J.Biol.Chem., 298, 2021
|
|
7N41
 
 | | Crystal structure of TagA with UDP-ManNAc | | Descriptor: | (2R,3S,4R,5S,6R)-3-acetamido-4,5-dihydroxy-6-(hydroxymethyl)oxan-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen diphosphate (non-preferred name), N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase | | Authors: | Martinez, O.E, Cascio, D, Clubb, R.T. | | Deposit date: | 2021-06-03 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Insight into the molecular basis of substrate recognition by the wall teichoic acid glycosyltransferase TagA.
J.Biol.Chem., 298, 2021
|
|
7CWQ
 
 | | Crystal structure of a novel cutinase from Burkhoderiales bacterium RIFCSPLOWO2_02_FULL_57_36 | | Descriptor: | DLH domain-containing protein, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2020-08-30 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | General features to enhance enzymatic activity of poly(ethylene terephthalate) hydrolysis.
Nat Catal, 4, 2021
|
|
