1YDX
 
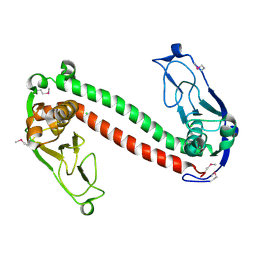 | | Crystal structure of Type-I restriction-modification system S subunit from M. genitalium | | Descriptor: | CHLORIDE ION, type I restriction enzyme specificity protein MG438 | | Authors: | Machado, B, Quijada, O, Pinol, J, Fita, I, Querol, E, Carpena, X. | | Deposit date: | 2004-12-27 | | Release date: | 2005-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Putative Type I Restriction-Modification S Subunit from Mycoplasma genitalium
J.Mol.Biol., 351, 2005
|
|
1GON
 
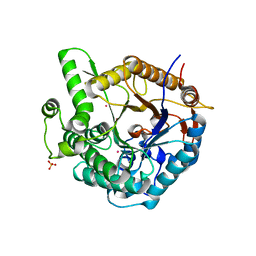 | | b-glucosidase from Streptomyces sp | | Descriptor: | BETA-GLUCOSIDASE, MERCURY (II) ION, SULFATE ION | | Authors: | Guasch, A, Perez-Pons, J.A, Vallmitjana, M, Querol, E, Coll, M. | | Deposit date: | 2001-10-22 | | Release date: | 2002-11-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Family 1 Beta-Glucosidase from Streptomyces
To be Published
|
|
1GNX
 
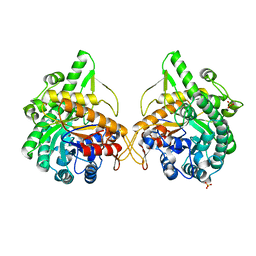 | | b-glucosidase from Streptomyces sp | | Descriptor: | BETA-GLUCOSIDASE, SULFATE ION, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Guasch, A, Perez-Pons, J.A, Vallmitjana, M, Querol, E, Coll, M. | | Deposit date: | 2001-10-10 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Beta-Glucosidase from Streptomyces
To be Published
|
|
4AMU
 
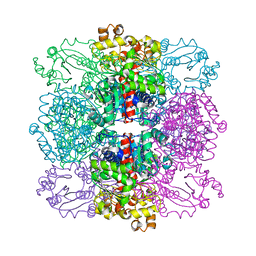 | | Structure of ornithine carbamoyltransferase from Mycoplasma penetrans with a P321 space group | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | Gallego, P, Benach, J, Planell, R, Querol, E, Perez-Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-13 | | Release date: | 2012-03-28 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
4ANF
 
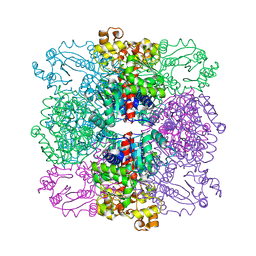 | | Structure of the ornithine carbamoyltransferase from Mycoplasma penetrans with a P23 Space group | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | Gallego, P, Benach, J, Planell, R, Querol, E, Perez-Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-16 | | Release date: | 2012-03-28 | | Last modified: | 2012-10-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
4AXS
 
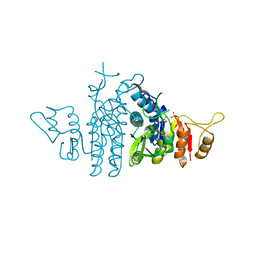 | | Structure of Carbamate Kinase from Mycoplasma penetrans | | Descriptor: | CARBAMATE KINASE, SULFATE ION | | Authors: | Gallego, P, Planell, R, Benach, J, Querol, E, PerezPons, J.A, Reverter, D. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma Penetrans.
Plos One, 7, 2012
|
|
4E4J
 
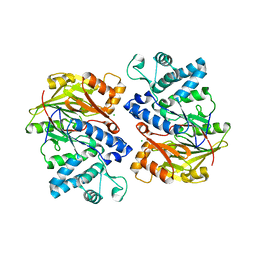 | | Crystal structure of arginine deiminase from Mycoplasma penetrans | | Descriptor: | Arginine deiminase, CHLORIDE ION | | Authors: | Benach, J, Gallego, P, Planell, R, Querol, E, Perez Pons, J.A, Reverter, D. | | Deposit date: | 2012-03-13 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of the Enzymes Composing the Arginine Deiminase Pathway in Mycoplasma penetrans.
Plos One, 7, 2012
|
|
1GBG
 
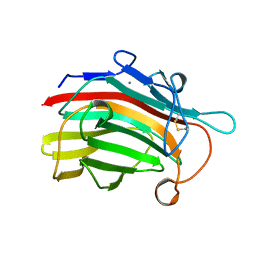 | | BACILLUS LICHENIFORMIS BETA-GLUCANASE | | Descriptor: | (1,3-1,4)-BETA-D-GLUCAN 4 GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase at 1.8 A resolution.
FEBS Lett., 374, 1995
|
|
5OX7
 
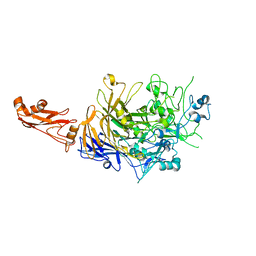 | |
4XNG
 
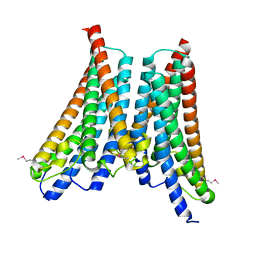 | |
6YRK
 
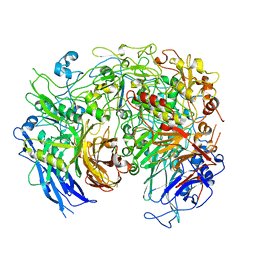 | |
4DCZ
 
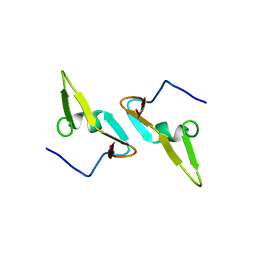 | |
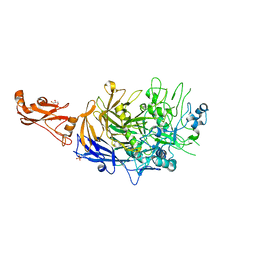
6R41
 
 | |
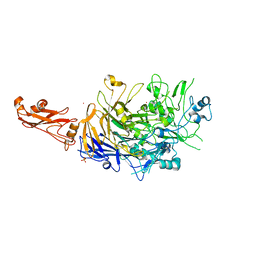
6R3T
 
 | |
6R43
 
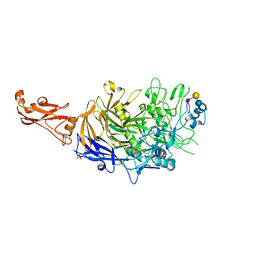 | | Structure of P110 from Mycoplasma Genitalium complexed with 6'-SL | | Descriptor: | Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Aparicio, D, Fita, I. | | Deposit date: | 2019-03-21 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Mycoplasma genitalium adhesin P110 binds sialic-acid human receptors.
Nat Commun, 9, 2018
|
|
6S3U
 
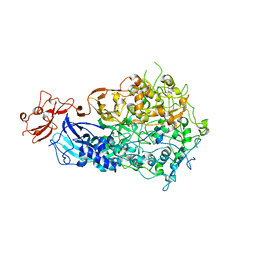 | | Adhesin P140 from Mycoplasma Genitalium | | Descriptor: | Adhesin P1 | | Authors: | Fita, I, Aparicio, D. | | Deposit date: | 2019-06-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structure and mechanism of the Nap adhesion complex from the human pathogen Mycoplasma genitalium.
Nat Commun, 11, 2020
|
|
6RUT
 
 | |
3D6E
 
 | | Crystal structure of the engineered 1,3-1,4-beta-glucanase protein from Bacillus licheniformis | | Descriptor: | Beta-glucanase, CALCIUM ION | | Authors: | Fita, I, Planas, A, Calisto, B.M, Addington, T. | | Deposit date: | 2008-05-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Re-engineering specificity in 1,3-1,4-beta-glucanase to accept branched xyloglucan substrates
Proteins, 79, 2011
|
|
