8K8H
 
 | |
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
7WYB
 
 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
7WY5
 
 | | ADGRL3/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7X10
 
 | | ADGRL3/miniG12 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
1S6C
 
 | | Crystal structure of the complex between KChIP1 and Kv4.2 N1-30 | | Descriptor: | CALCIUM ION, Kv4 potassium channel-interacting protein KChIP1b, Potassium voltage-gated channel subfamily D member 2 | | Authors: | Zhou, W, Qian, Y, Kunjilwar, K, Pfaffinger, P.J, Choe, S. | | Deposit date: | 2004-01-23 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the functional interaction of KChIP1 with Shal-type K(+) channels.
Neuron, 41, 2004
|
|
4GQB
 
 | | Crystal Structure of the human PRMT5:MEP50 Complex | | Descriptor: | (2S,5S,6E)-2,5-diamino-6-[(3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxydihydrofuran-2(3H)-ylidene]hexanoic acid, Histone H4 peptide, Methylosome protein 50, ... | | Authors: | Antonysamy, S, Bonday, Z, Campbell, R, Doyle, B, Druzina, Z, Gheyi, T, Han, B, Jungheim, L.N, Qian, Y, Rauch, C, Russell, M, Sauder, J.M, Wasserman, S.R, Weichert, K, Willard, F.S, Zhang, A, Emtage, S. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-17 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the human PRMT5:MEP50 complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4I5I
 
 | | Crystal structure of the SIRT1 catalytic domain bound to NAD and an EX527 analog | | Descriptor: | (6S)-2-chloro-5,6,7,8,9,10-hexahydrocyclohepta[b]indole-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Zhao, X, Allison, D, Condon, B, Zhang, F, Gheyi, T, Zhang, A, Ashok, S, Russell, M, Macewan, I, Qian, Y, Jamison, J.A, Luz, J.G. | | Deposit date: | 2012-11-28 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 2.5 angstrom crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition.
J.Med.Chem., 56, 2013
|
|
4M5E
 
 | | Tse3 structure | | Descriptor: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Qian, Y. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
5ZV7
 
 | | P domain of GII.17-2014/15 complexed with B-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZVC
 
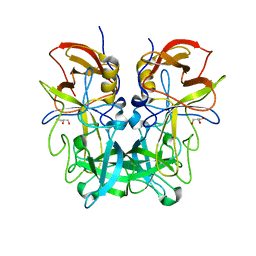 | | P domain of GII.13 norovirus capsid complexed with Lewis A trisaccharide | | Descriptor: | GLYCEROL, Major capsid protein VP1, beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZV9
 
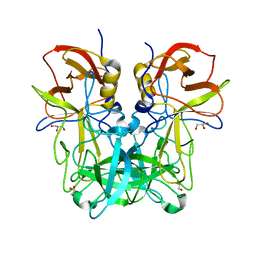 | | P domain of GII.13 norovirus capsid | | Descriptor: | GLYCEROL, Major capsid protein VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZUQ
 
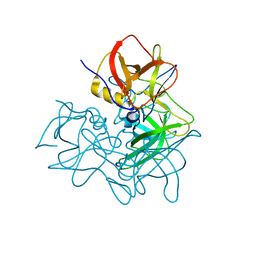 | | P domain of GII.17-1978 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZUS
 
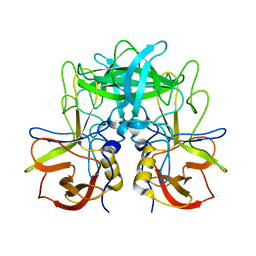 | | P domain of GII.17-2014/15 | | Descriptor: | VP1 | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
5ZV5
 
 | | P domain of GII.17-2014/15 complexed with A-trisaccharide | | Descriptor: | VP1, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]alpha-D-galactopyranose | | Authors: | Chen, Y, Li, X. | | Deposit date: | 2018-05-09 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Adaptations of Norovirus GII.17/13/21 Lineage through Two Distinct Evolutionary Paths.
J. Virol., 93, 2019
|
|
7WCM
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist MBX-2982 | | Descriptor: | 2-[1-(5-ethylpyrimidin-2-yl)piperidin-4-yl]-4-[[4-(1,2,3,4-tetrazol-1-yl)phenoxy]methyl]-1,3-thiazole, Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
7WCN
 
 | | Cryo-EM structure of GPR119-Gs Complex with small molecule agonist AR231453 | | Descriptor: | Glucose-dependent insulinotropic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qiao, A.N, Wu, S, Ye, S. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-21 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Activation and signaling mechanism revealed by GPR119-G s complex structures.
Nat Commun, 13, 2022
|
|
4R6U
 
 | | IL-18 receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-18, Interleukin-18 receptor 1 | | Authors: | Wei, H, Wang, X. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the specific recognition of IL-18 by its alpha receptor.
Febs Lett., 588, 2014
|
|
8WW2
 
 | | GPR3/Gs complex | | Descriptor: | CHOLESTEROL HEMISUCCINATE, G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xiong, Y. | | Deposit date: | 2023-10-24 | | Release date: | 2024-02-14 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Identification of oleic acid as an endogenous ligand of GPR3.
Cell Res., 34, 2024
|
|
8HBC
 
 | | Crystal structure of the CysR-CTLD3 fragment of human DEC205 | | Descriptor: | Lymphocyte antigen 75 | | Authors: | Kong, D, Yu, B, Hu, Z, Cheng, C, Cao, L, He, Y. | | Deposit date: | 2022-10-28 | | Release date: | 2023-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Interaction of human dendritic cell receptor DEC205/CD205 with keratins.
J.Biol.Chem., 300, 2024
|
|
1HOM
 
 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Qian, Y.-Q, Billeter, M, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
5UKG
 
 | |
6PX2
 
 | | Acropora millepora GAPDH | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Brandt, G.S, Fields, P.A. | | Deposit date: | 2019-07-24 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thermal stability and structure of glyceraldehyde-3-phosphate dehydrogenase from the coral Acropora millepora.
Rsc Adv, 11, 2021
|
|
8KAE
 
 | | 16d-bound human SPNS2 | | Descriptor: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, NbFab chain L, NbFab-H chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
5HFU
 
 | | Crystal Structure of Human Hexokinase 2 with cmpd 27, a 2-amido-6-benzenesulfonamide glucosamine | | Descriptor: | Hexokinase-2, ~{N}-[(2~{S},3~{R},4~{R},5~{S},6~{R})-6-[[(4-cyanophenyl)sulfonylamino]methyl]-2,4,5-tris(oxidanyl)oxan-3-yl]-3-phenyl-benzamide | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.923 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
