6AZ0
 
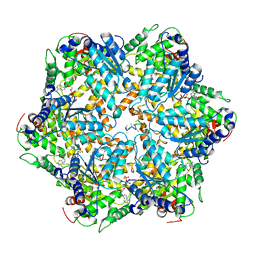 | | Mitochondrial ATPase Protease YME1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Puchades, C, Rampello, A.J, Shin, M, Giuliano, C, Wiseman, R.L, Glynn, S.E, Lander, G.C. | | Deposit date: | 2017-09-09 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing.
Science, 358, 2017
|
|
6NYY
 
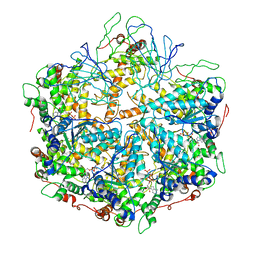 | | human m-AAA protease AFG3L2, substrate-bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AFG3-like protein 2, MAGNESIUM ION, ... | | Authors: | Lander, G.C, Puchades, C. | | Deposit date: | 2019-02-12 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unique Structural Features of the Mitochondrial AAA+ Protease AFG3L2 Reveal the Molecular Basis for Activity in Health and Disease.
Mol.Cell, 75, 2019
|
|
6V11
 
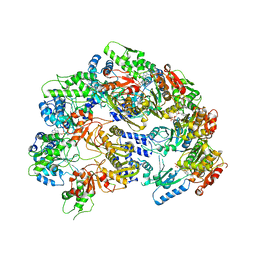 | | Lon Protease from Yersinia pestis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Shin, M, Puchades, C, Asmita, A, Puri, N, Adjei, E, Wiseman, R.L, Karzai, A.W, Lander, G.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Sci Adv, 6, 2020
|
|
6ON2
 
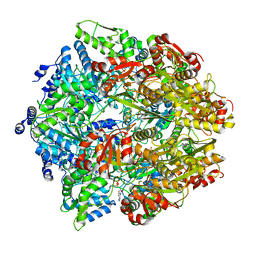 | | Lon Protease from Yersinia pestis with Y2853 substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent protease La, ... | | Authors: | Shin, M, Asmita, A, Puchades, C, Adjei, E, Wiseman, R.L, Karzai, A.W, Lander, G.C. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Sci Adv, 6, 2020
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KDT
 
 | |
8SUR
 
 | | TMEM16F bound with Niclosamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloro-N-(2-chloro-4-nitrophenyl)-2-hydroxybenzamide, ... | | Authors: | Feng, S, Cheng, Y. | | Deposit date: | 2023-05-13 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8TAG
 
 | | TMEM16F, with Calcium and PIP2, no inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Cheng, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8TAL
 
 | | TMEM16F, with Calcium and PIP2, no inhibitor, Cl1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | Authors: | Wu, H, Feng, S, Cheng, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8TAI
 
 | | TMEM16F, with Calcium and PIP2, no inhibitor, Cl2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | Authors: | Wu, H, Feng, S, Cheng, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
8SUN
 
 | | TMEM16F 1PBC | | Descriptor: | 1-Hydroxy-3-(trifluoromethyl)pyrido[1,2-a]benzimidazole-4-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, ... | | Authors: | Wu, H, Feng, S, Cheng, Y. | | Deposit date: | 2023-05-12 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
5CEZ
 
 | |
5CEY
 
 | |
5CEX
 
 | |
