6QM3
 
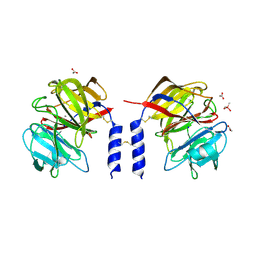 | | Crystal structure of a calcium- and sodium-bound mouse Olfactomedin-1 disulfide-linked dimer of the Olfactomedin domain and part of coiled coil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CACODYLATE ION, ... | | Authors: | Pronker, M.F, van den Hoek, H.G, Janssen, B.J.C. | | Deposit date: | 2019-02-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol, 20, 2019
|
|
5AMO
 
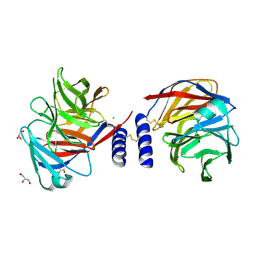 | | Structure of a mouse Olfactomedin-1 disulfide-linked dimer of the Olfactomedin domain and part of the coiled coil | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Pronker, M.F, Bos, T.G.A.A, Sharp, T.H, Thies-Weesie, D.M, Janssen, B.J.C. | | Deposit date: | 2015-03-11 | | Release date: | 2015-04-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Olfactomedin-1 Has a V-Shaped Disulfide-Linked Tetrameric Structure.
J.Biol.Chem., 290, 2015
|
|
6QHJ
 
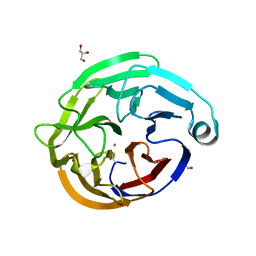 | | High-resolution crystal structure of calcium- and sodium-bound mouse Olfactomedin-1 beta-propeller domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Pronker, M.F, van den Hoek, H.G, Janssen, B.J.C. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol, 20, 2019
|
|
5LFR
 
 | |
5LFV
 
 | |
5LF5
 
 | |
5LFU
 
 | |
8OPN
 
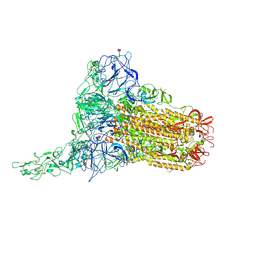 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (1-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OHN
 
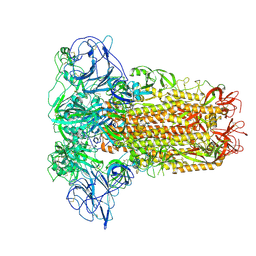 | | Human Coronavirus HKU1 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pronker, M.F, Hurdiss, D.L. | | Deposit date: | 2023-03-21 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPO
 
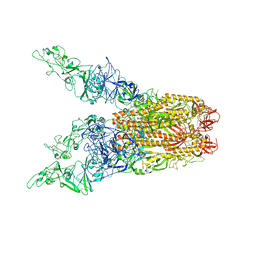 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (3-up state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
8OPM
 
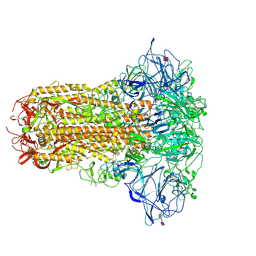 | | Human Coronavirus HKU1 spike glycoprotein in complex with an alpha2,8-linked 9-O-acetylated disialoside (closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-O-acetyl-5-acetamido-3,5-dideoxy-D-glycero-alpha-D-galacto-non-2-ulopyranosonic acid-(2-8)-N-acetyl-alpha-neuraminic acid, ... | | Authors: | Pronker, M.F, Creutznacher, R, Hurdiss, D.L. | | Deposit date: | 2023-04-07 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Sialoglycan binding triggers spike opening in a human coronavirus.
Nature, 624, 2023
|
|
5O0K
 
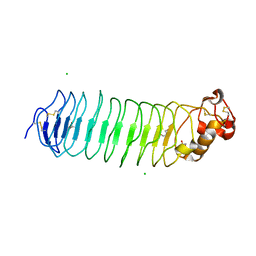 | |
5O0M
 
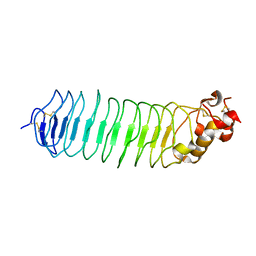 | |
5O0R
 
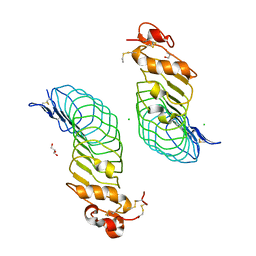 | |
5O0O
 
 | |
5O0L
 
 | |
5O0P
 
 | |
5O0N
 
 | | Deglycosylated Nogo Receptor with native disulfide structure 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Reticulon-4 receptor, ... | | Authors: | Pronker, M.F, Tas, R.P, Vlieg, H.C, Janssen, B.J.C. | | Deposit date: | 2017-05-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nogo Receptor crystal structures with a native disulfide pattern suggest a novel mode of self-interaction.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5O0Q
 
 | |
7BG1
 
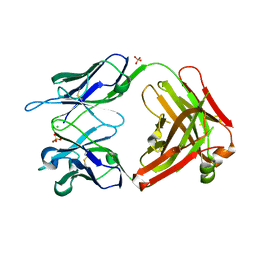 | |
8RMO
 
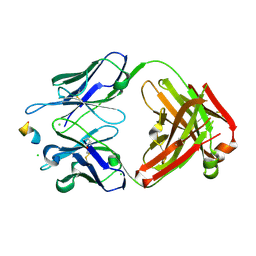 | |
6Y3Y
 
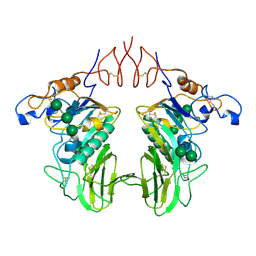 | | Human Coronavirus HKU1 Haemagglutinin-Esterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin-esterase, ... | | Authors: | Hurdiss, D.L, Drulyte, I, Pronker, M.F. | | Deposit date: | 2020-02-19 | | Release date: | 2020-04-08 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structure of coronavirus-HKU1 haemagglutinin esterase reveals architectural changes arising from prolonged circulation in humans.
Nat Commun, 11, 2020
|
|
8P6I
 
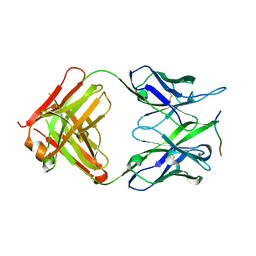 | | Crystal structure of the 139H2 Fab fragment bound to Muc1 peptide epitope | | Descriptor: | 139H2 HC, 139H2 LC, Mucin-1 | | Authors: | Beugelink, J.W, Peng, W, Siborova, M, Pronker, M.F, Snijder, J, Janssen, B.J.C. | | Deposit date: | 2023-05-26 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reverse-engineering the anti-MUC1 antibody 139H2 by mass spectrometry-based de novo sequencing.
Life Sci Alliance, 7, 2024
|
|
