3TB6
 
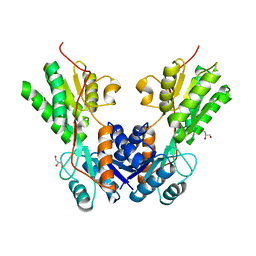 | |
3FZY
 
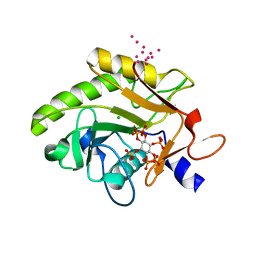 | | Crystal Structure of Pre-cleavage Form of Cysteine Protease Domain from Vibrio cholerae RtxA Toxin | | Descriptor: | CHLORIDE ION, INOSITOL HEXAKISPHOSPHATE, RTX toxin RtxA, ... | | Authors: | Shuvalova, L, Minasov, G, Prochazkova, K, Satchell, K.J.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-01-26 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and molecular mechanism for autoprocessing of MARTX toxin of Vibrio cholerae at multiple sites
J.Biol.Chem., 284, 2009
|
|
2N3A
 
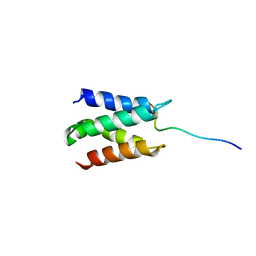 | | Solution structure of LEDGF/p75 IBD in complex with POGZ peptide (1389-1404) | | Descriptor: | PC4 and SFRS1-interacting protein, Pogo transposable element with ZNF domain | | Authors: | Tesina, P, Cermakova, K, Horejsi, M, Prochazkova, K, Fabry, M, Sharma, S, Christ, F, Demeulemeester, J, Debyser, Z, De Rijck, J, Veverka, V, Rezacova, P. | | Deposit date: | 2015-05-26 | | Release date: | 2015-08-19 | | Method: | SOLUTION NMR | | Cite: | Multiple cellular proteins interact with LEDGF/p75 through a conserved unstructured consensus motif.
Nat Commun, 6, 2015
|
|
3CJB
 
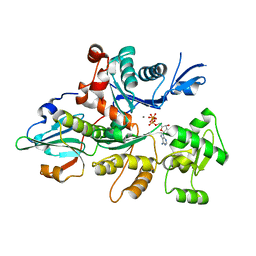 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with Gelsolin-segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3CJC
 
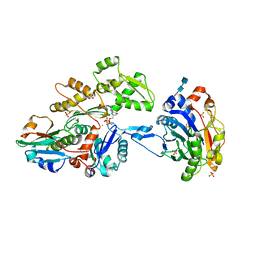 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with DNase I and Gelsolin-segment 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5EDF
 
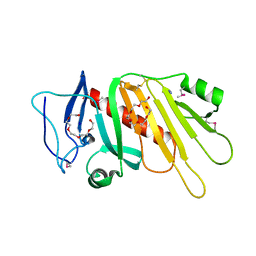 | | Crystal structure of the selenomethionine-substituted iron-regulated protein FrpD from Neisseria meningitidis | | Descriptor: | AZIDE ION, FrpC operon protein, HEXAETHYLENE GLYCOL, ... | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
5EDJ
 
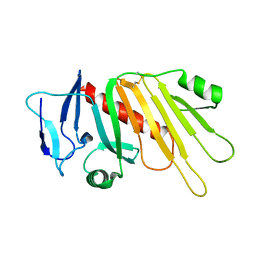 | | Crystal structure of the Neisseria meningitidis iron-regulated outer membrane lipoprotein FrpD | | Descriptor: | FrpC operon protein | | Authors: | Sviridova, E, Bumba, L, Rezacova, P, Sebo, P, Kuta Smatanova, I. | | Deposit date: | 2015-10-21 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of the interaction between the putative adhesion-involved and iron-regulated FrpD and FrpC proteins of Neisseria meningitidis.
Sci Rep, 7, 2017
|
|
4OQP
 
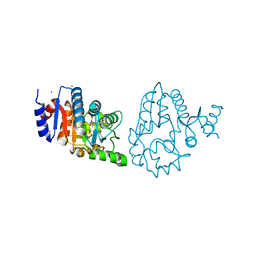 | |
4OQQ
 
 | |
