8EC6
 
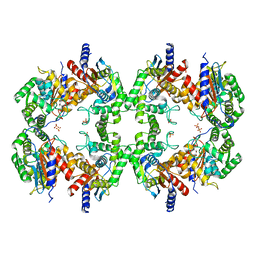 | | Cryo-EM structure of the Glutaminase C core filament (fGAC) | | Descriptor: | Isoform 2 of Glutaminase kidney isoform, mitochondrial, PHOSPHATE ION | | Authors: | Ambrosio, A.L, Dias, S.M, Quesnay, J.E, Portugal, R.V, Cassago, A, van Heel, M.G, Islam, Z, Rodrigues, C.T. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular mechanism of glutaminase activation through filamentation and the role of filaments in mitophagy protection.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6N4C
 
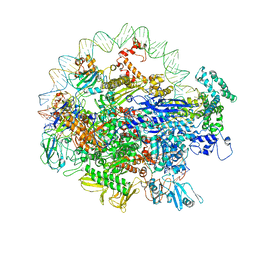 | | EM structure of the DNA wrapping in bacterial open transcription initiation complex | | Descriptor: | DNA (94-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Florez-Ariza, A, Cassago, A, de Oliveira, P.S.L, Guerra, D.G, Portugal, R.V. | | Deposit date: | 2018-11-19 | | Release date: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Interactions of Upstream and Downstream Promoter Regions with RNA Polymerase are Energetically Coupled and a Target of Regulation in Transcription Initiation
Biorxiv, 2020
|
|
7KO8
 
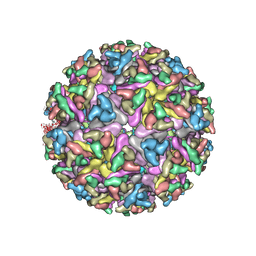 | | Cryo-EM structure of the mature and infective Mayaro virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein, E1 glycoprotein, ... | | Authors: | Riberio-Filho, H.V, Coimbra, L.D, Cassago, A, Rocha, R.P.F, Padilha, A.C.M, Schatz, M, van Heel, M.G, Portugal, R.V, Trivella, D.B.B, de Oliveira, P.S.L, Marques, R.E. | | Deposit date: | 2020-11-06 | | Release date: | 2021-04-28 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the mature and infective Mayaro virus at 4.4 angstrom resolution reveals features of arthritogenic alphaviruses.
Nat Commun, 12, 2021
|
|
7UFR
 
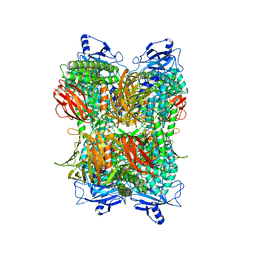 | | Cryo-EM Structure of Bl_Man38A at 2.7 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structure of Bl_Man38A at 2.7 A
Nat.Chem.Biol., 2022
|
|
7UFU
 
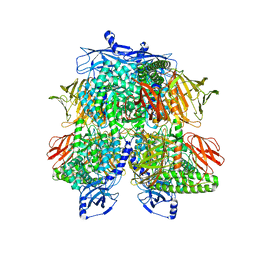 | | Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A | | Descriptor: | Alpha-mannosidase, ZINC ION, alpha-D-mannopyranose | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM Structure of Bl_Man38A nucleophile mutant in complex with mannose at 2.7 A
Nat.Chem.Biol., 2022
|
|
7UFT
 
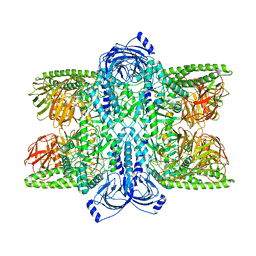 | | Cryo-EM Structure of Bl_Man38C at 2.9 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structure of Bl_Man38C at 2.9 A
Nat.Chem.Biol., 2022
|
|
7UFS
 
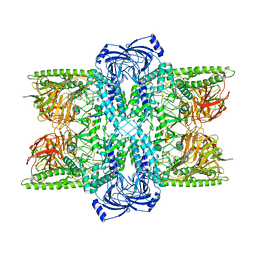 | | Cryo-EM Structure of Bl_Man38B at 3.4 A | | Descriptor: | Alpha-mannosidase, ZINC ION | | Authors: | Santos, C.R, Cordeiro, R.L, Domingues, M.N, Borges, A.C, de Farias, M.A, Van Heel, M, Murakami, M.T, Portugal, R.V. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM Structure of Bl_Man38B at 3.4 A
Nat.Chem.Biol., 2022
|
|
7M6J
 
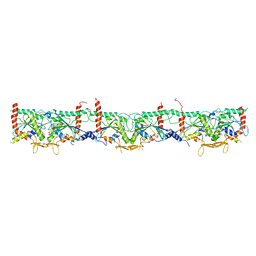 | | Human Septin Hexameric Complex SEPT2G/SEPT6/SEPT7 by Single Particle Cryo-EM | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mendonca, D.C, Pereira, H.M, van Heel, M, Portugal, R.V, Garratt, R.C. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An atomic model for the human septin hexamer by cryo-EM.
J.Mol.Biol., 433, 2021
|
|
5M3L
 
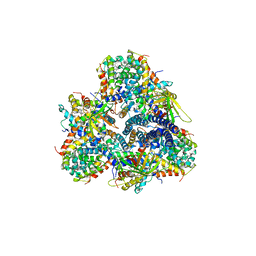 | | Single-particle cryo-EM using alignment by classification (ABC): the structure of Lumbricus terrestris hemoglobin | | Descriptor: | Extracellular globin-2, Extracellular globin-3, Extracellular globin-4, ... | | Authors: | Afanasyev, P, Linnemayr-Seer, C, Ravelli, R.B.G, Matadeen, R, De Carlo, S, Alewijnse, B, Portugal, R.V, Pannu, N.S, Schatz, M, van Heel, M. | | Deposit date: | 2016-10-15 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Single-particle cryo-EM using alignment by classification (ABC): the structure of Lumbricus terrestris haemoglobin.
IUCrJ, 4, 2017
|
|
4JKT
 
 | | Crystal structure of mouse Glutaminase C, BPTES-bound form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Fornezari, C, Ferreira, A.P.S, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2013-03-11 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Active Glutaminase C Self-assembles into a Supratetrameric Oligomer That Can Be Disrupted by an Allosteric Inhibitor.
J.Biol.Chem., 288, 2013
|
|
3SUK
 
 | | Crystal structure of cerato-platanin 2 from M. perniciosa (MpCP2) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUJ
 
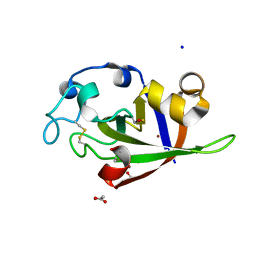 | | Crystal structure of cerato-platanin 1 from M. perniciosa (MpCP1) | | Descriptor: | ACETATE ION, CHLORIDE ION, Cerato-platanin 1, ... | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUM
 
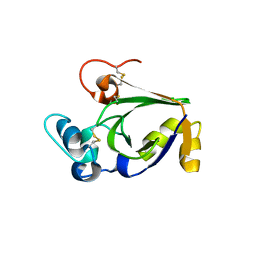 | | Crystal structure of cerato-platanin 5 from M. perniciosa (MpCP5) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
3SUL
 
 | | Crystal structure of cerato-platanin 3 from M. perniciosa (MpCP3) | | Descriptor: | Cerato-platanin-like protein | | Authors: | Oliveira, J.F, Barsottini, M.R.O, Zaparoli, G, Machado, L.O, Dias, S.M.G, Pereira, G.A.G, Ambrosio, A.L.B. | | Deposit date: | 2011-07-11 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Functional diversification of cerato-platanins in Moniliophthora perniciosa as seen by differential expression and protein function specialization.
Mol. Plant Microbe Interact., 26, 2013
|
|
6GYB
 
 | | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri | | Descriptor: | VirB10 protein, VirB7, VirB9 protein | | Authors: | Sgro, G.G, Costa, T.R.D, Farah, C.S, Waksman, G. | | Deposit date: | 2018-06-28 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri.
Nat Microbiol, 3, 2018
|
|
5U0K
 
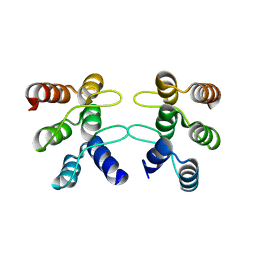 | | C-terminal ankyrin repeats from human liver-type glutaminase (GAB/LGA) | | Descriptor: | Glutaminase liver isoform, mitochondrial | | Authors: | Ferreira, I.M, Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.548 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5U0J
 
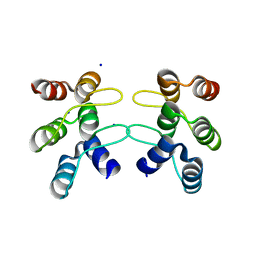 | | C-terminal ankyrin repeats from human kidney-type glutaminase (KGA) - monoclinic crystal form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, SODIUM ION | | Authors: | Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5U0I
 
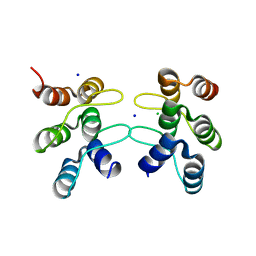 | | C-terminal ankyrin repeats from human kidney-type glutaminase (KGA) - tetragonal crystal form | | Descriptor: | CHLORIDE ION, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Pasquali, C.C, Gonzalez, A, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
5UQE
 
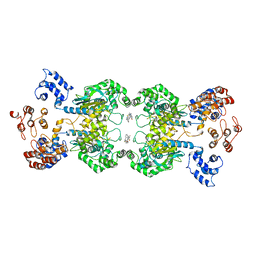 | | Multidomain structure of human kidney-type glutaminase(KGA/GLS) | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Pasquali, C.C, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2017-02-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
