2LSM
 
 | | Solution structure of gpFI C-terminal domain | | Descriptor: | DNA-packaging protein FI | | Authors: | Popovic, A, Wu, B, Edwards, A.M, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization of phage lambda FI protein (gpFI) reveals a novel mechanism of DNA packaging chaperone activity.
J.Biol.Chem., 287, 2012
|
|
4V7G
 
 | | Crystal Structure of Lumazine Synthase from Bacillus Anthracis | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Morgunova, E, Illarionov, B, Saller, S, Popov, A, Sambaiah, T, Bacher, A, Cushman, M, Fischer, M, Ladenstein, R. | | Deposit date: | 2009-09-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study and thermodynamic characterization of inhibitor binding to lumazine synthase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4XA7
 
 | | Soluble part of holo NqrC from V. harveyi | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit C | | Authors: | Borshchevskiy, V, Round, E, Bertsova, Y, Polovinkin, V, Gushchin, I, Mishin, A, Kovalev, K, Kachalova, G, Popov, A, Bogachev, A, Gordeliy, V. | | Deposit date: | 2014-12-12 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Functional Investigation of Flavin Binding Center of the NqrC Subunit of Sodium-Translocating NADH:Quinone Oxidoreductase from Vibrio harveyi.
Plos One, 10, 2015
|
|
5TJE
 
 | | Murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 and T cell receptor P14 | | Descriptor: | ALPHA CHAIN OF MURINE T CELL RECEPTOR p14, BETA CHAIN OF MURINE T CELL RECEPTOR p14, Beta-2-microglobulin, ... | | Authors: | Achour, A, Sandalova, T, Allerbring, E, Popov, A. | | Deposit date: | 2016-10-04 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
to be published
|
|
1USY
 
 | | ATP phosphoribosyl transferase (HisG:HisZ) complex from Thermotoga maritima | | Descriptor: | ATP PHOSPHORIBOSYLTRANSFERASE, ATP PHOSPHORIBOSYLTRANSFERASE REGULATORY SUBUNIT, HISTIDINE, ... | | Authors: | Vega, M.C, Fernandez, F.J, Murphy, G.E, Zou, P, Popov, A, Wilmanns, M. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-15 | | Last modified: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Regulation of the Hetero-Octameric ATP Phosphoribosyl Transferase Complex from Thermotoga Maritima by a tRNA Synthetase-Like Subunit.
Mol.Microbiol., 55, 2005
|
|
1YA5
 
 | | Crystal structure of the titin domains z1z2 in complex with telethonin | | Descriptor: | N2B-TITIN ISOFORM, SULFATE ION, TELETHONIN | | Authors: | Pinotsis, N, Popov, A, Zou, P, Wilmanns, M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-12-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.445 Å) | | Cite: | Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk
Nature, 439, 2006
|
|
5EGO
 
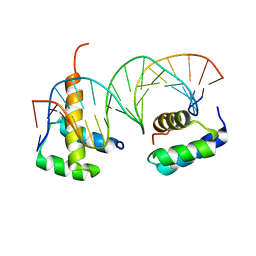 | | HOXB13-MEIS1 heterodimer bound to methylated DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*G)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
5EF6
 
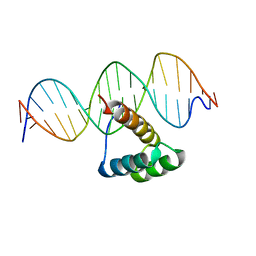 | | Structure of HOXB13 complex with methylated DNA | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*TP*(5CM)P*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*(5CM)P*GP*AP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-23 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
5EEA
 
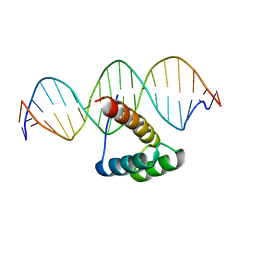 | | Structure of HOXB13-DNA(CAA) complex | | Descriptor: | DNA (5'-D(P*GP*GP*AP*CP*CP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*GP*GP*TP*CP*C)-3'), Homeobox protein Hox-B13 | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-22 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
5EG0
 
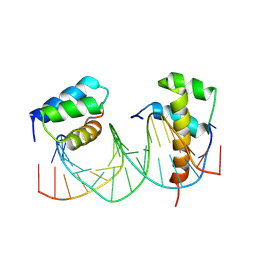 | | HOXB13-MEIS1 heterodimer bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*CP*GP*TP*AP*AP*AP*AP*CP*TP*GP*TP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*AP*CP*AP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*G)-3'), Homeobox protein Hox-B13, ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Molecular basis of recognition of two distinct DNA sequences by a single transcription factor
To Be Published
|
|
5EDN
 
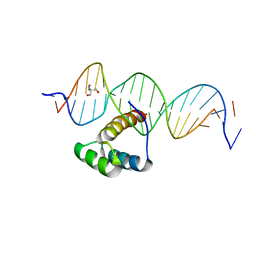 | | Structure of HOXB13-DNA(TCG) complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (5'-D(P*GP*GP*AP*CP*CP*TP*CP*GP*TP*AP*AP*AP*AP*CP*AP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*CP*GP*AP*GP*GP*TP*CP*C)-3'), ... | | Authors: | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | Deposit date: | 2015-10-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
5IJI
 
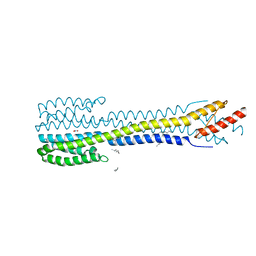 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in symmetric holo state | | Descriptor: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor histidine kinase NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-03-02 | | Release date: | 2017-05-31 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
5JEF
 
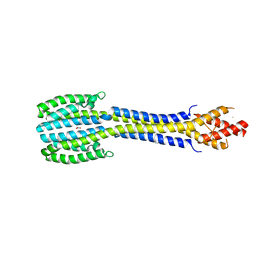 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in asymmetric holo state | | Descriptor: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
5JEQ
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50K) in symmetric apo state | | Descriptor: | Nitrate/nitrite sensor protein NarQ, PHOSPHATE ION | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
8OSB
 
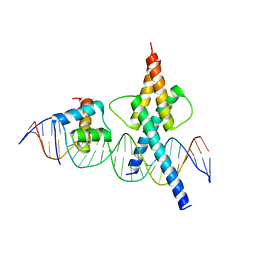 | | TWIST1-TCF4-ALX4 complex on specific DNA | | Descriptor: | DNA (25-MER), Homeobox protein aristaless-like 4, Transcription factor 4, ... | | Authors: | Morgunova, E, Kim, S, Popov, A, Wysocka, J, Taipale, J. | | Deposit date: | 2023-04-18 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | DNA-guided transcription factor cooperativity shapes face and limb mesenchyme.
Cell, 187, 2024
|
|
5JGP
 
 | | Crystal structure of the nitrate/nitrite sensor NarQ fragment bound with iodide ions | | Descriptor: | IODIDE ION, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | Authors: | Melnikov, I, Polovinkin, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
8BYX
 
 | |
8BZM
 
 | | FOXK1-ELF1-heterodimer bound to DNA | | Descriptor: | DNA, ETS-related transcription factor Elf-1, Forkhead box protein K1, ... | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | FOXK1-ELF1_heterodimer bound to DNA
To Be Published
|
|
5JTB
 
 | | Crystal structure of the chimeric protein of A2aAR-BRIL with bound iodide ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Melnikov, I, Polovinkin, V, Shevtsov, M, Borshchevskiy, V, Cherezov, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-09 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
5JRF
 
 | | Crystal structure of the light-driven sodium pump KR2 bound with iodide ions | | Descriptor: | EICOSANE, IODIDE ION, Sodium pumping rhodopsin | | Authors: | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-06 | | Release date: | 2017-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
5JSI
 
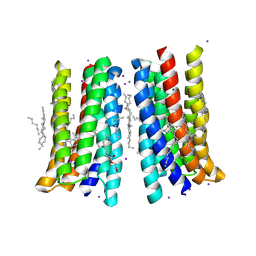 | | Structure of membrane protein | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, EICOSANE, ... | | Authors: | Melnikov, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gushchin, I, Popov, A, Gordeliy, V. | | Deposit date: | 2016-05-08 | | Release date: | 2017-05-31 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
4LU1
 
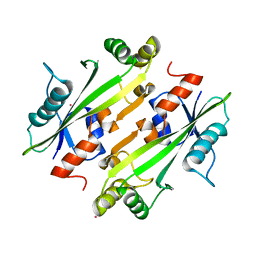 | | Crystal structure of the uncharacterized Maf protein YceF from E. coli, mutant D69A | | Descriptor: | Maf-like protein YceF, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Xu, X, Cui, H, Tchigvintsev, A, Flick, R, Brown, G, Popovic, A, Yakunin, A.F, Savchenko, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
4NY6
 
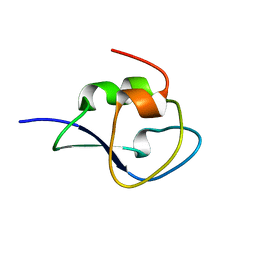 | | Neutron structure of leucine and valine methyl protonated type III antifreeze | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Fisher, S.J, Blakeley, M.P, Howard, E.I, Petite-Haertlein, I, Haertlein, M, Mitschler, A, Cousido-Siah, A, Salvaya, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A.D. | | Deposit date: | 2013-12-10 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.05 Å), X-RAY DIFFRACTION | | Cite: | Perdeuteration: improved visualization of solvent structure in neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5D8M
 
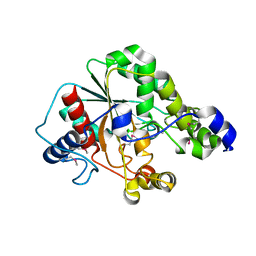 | | Crystal structure of the metagenomic carboxyl esterase MGS0156 | | Descriptor: | Metagenomic carboxyl esterase MGS0156 | | Authors: | Cui, H, Nocek, B, Tchigvintsev, A, Popovic, A, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2015-08-17 | | Release date: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of esterase (MGS0156)
To Be Published
|
|
4E6B
 
 | |
