1CDK
 
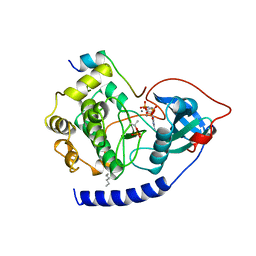 | | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT (E.C.2.7.1.37) (PROTEIN KINASE A) COMPLEXED WITH PROTEIN KINASE INHIBITOR PEPTIDE FRAGMENT 5-24 (PKI(5-24) ISOELECTRIC VARIANT CA) AND MN2+ ADENYLYL IMIDODIPHOSPHATE (MNAMP-PNP) AT PH 5.6 AND 7C AND 4C | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, MANGANESE (II) ION, MYRISTIC ACID, ... | | Authors: | Bossemeyer, D, Engh, R.A, Kinzel, V, Ponstingl, H, Huber, R. | | Deposit date: | 1994-07-04 | | Release date: | 1995-10-15 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24).
EMBO J., 12, 1993
|
|
3GRX
 
 | | NMR STRUCTURE OF ESCHERICHIA COLI GLUTAREDOXIN 3-GLUTATHIONE MIXED DISULFIDE COMPLEX, 20 STRUCTURES | | Descriptor: | GLUTAREDOXIN 3, GLUTATHIONE | | Authors: | Nordstrand, K, Aslund, F, Holmgren, A, Otting, G, Berndt, K.D. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-30 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism.
J.Mol.Biol., 286, 1999
|
|
1MPH
 
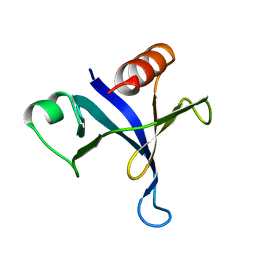 | | PLECKSTRIN HOMOLOGY DOMAIN FROM MOUSE BETA-SPECTRIN, NMR, 50 STRUCTURES | | Descriptor: | BETA SPECTRIN | | Authors: | Nilges, M, Macias, M.J, O'Donoghue, S.I, Oschkinat, H. | | Deposit date: | 1997-04-23 | | Release date: | 1997-06-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Automated NOESY interpretation with ambiguous distance restraints: the refined NMR solution structure of the pleckstrin homology domain from beta-spectrin.
J.Mol.Biol., 269, 1997
|
|
1YDT
 
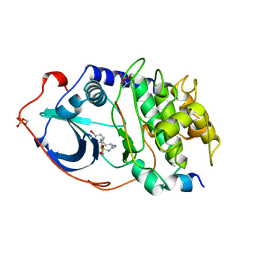 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDS
 
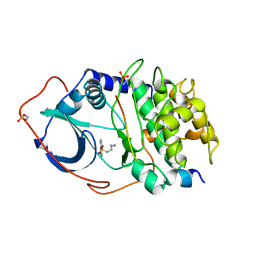 | | Structure of CAMP-dependent protein kinase, alpha-catalytic subunit in complex with H8 protein kinase inhibitor [N-(2-methylamino)ethyl]-5-isoquinolinesulfonamide | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(METHYLAMINO)ETHYL]-5-ISOQUINOLINESULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2012-02-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDR
 
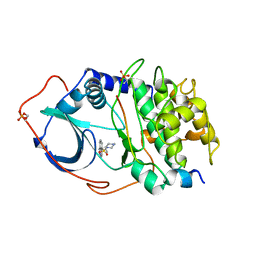 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE | | Descriptor: | 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE, C-AMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1RRP
 
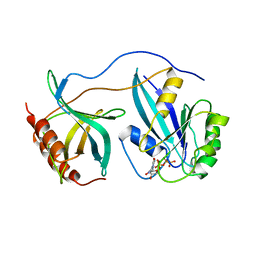 | | STRUCTURE OF THE RAN-GPPNHP-RANBD1 COMPLEX | | Descriptor: | MAGNESIUM ION, NUCLEAR PORE COMPLEX PROTEIN NUP358, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Vetter, I.R, Nowak, C, Nishimoto, T, Kuhlmann, J, Wittinghofer, A. | | Deposit date: | 1999-01-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport.
Nature, 398, 1999
|
|
1STC
 
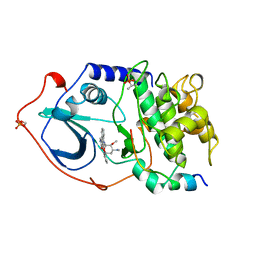 | | CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR, STAUROSPORINE | | Authors: | Prade, L, Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1997-10-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential.
Structure, 5, 1997
|
|
1K5G
 
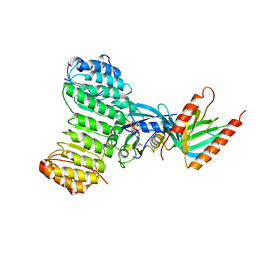 | | Crystal structure of Ran-GDP-AlFx-RanBP1-RanGAP complex | | Descriptor: | ALUMINUM FLUORIDE, GTP-binding nuclear protein RAN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
1K5D
 
 | | Crystal structure of Ran-GPPNHP-RanBP1-RanGAP complex | | Descriptor: | GTP-binding nuclear protein RAN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Seewald, M.J, Koerner, C, Wittinghofer, A, Vetter, I.R. | | Deposit date: | 2001-10-10 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RanGAP mediates GTP hydrolysis without an arginine finger.
Nature, 415, 2002
|
|
1YRG
 
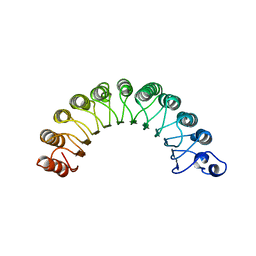 | | THE CRYSTAL STRUCTURE OF RNA1P: A NEW FOLD FOR A GTPASE-ACTIVATING PROTEIN | | Descriptor: | GTPASE-ACTIVATING PROTEIN RNA1_SCHPO | | Authors: | Hillig, R.C, Renault, L, Vetter, I.R, Drell, T, Wittinghofer, A, Becker, J. | | Deposit date: | 1999-03-29 | | Release date: | 2000-03-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | The crystal structure of rna1p: a new fold for a GTPase-activating protein.
Mol.Cell, 3, 1999
|
|
1Q61
 
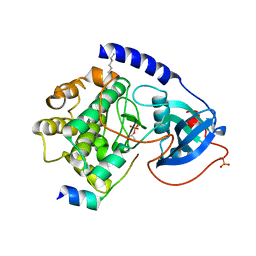 | | PKA triple mutant model of PKB | | Descriptor: | N-OCTANOYL-N-METHYLGLUCAMINE, cAMP-dependent protein kinase inhibitor, alpha form, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-09-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1Q62
 
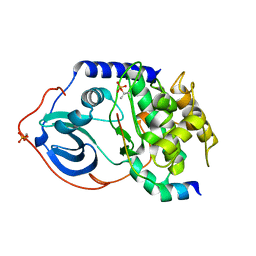 | | PKA double mutant model of PKB | | Descriptor: | cAMP-dependent protein kinase inhibitor, alpha form, cAMP-dependent protein kinase, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-08-12 | | Release date: | 2003-09-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
1Q24
 
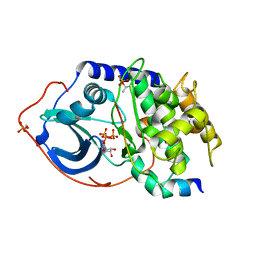 | | PKA double mutant model of PKB in complex with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, cAMP-dependent Protein Kinase Inhibitor, ... | | Authors: | Gassel, M, Breitenlechner, C.B, Rueger, P, Jucknischke, U, Schneider, T, Huber, R, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2003-07-23 | | Release date: | 2003-08-19 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of protein kinase B (AKT)
J.Mol.Biol., 329, 2003
|
|
