1C8B
 
 | | CRYSTAL STRUCTURE OF A NOVEL GERMINATION PROTEASE FROM SPORES OF BACILLUS MEGATERIUM: STRUCTURAL REARRANGEMENTS AND ZYMOGEN ACTIVATION | | Descriptor: | SPORE PROTEASE | | Authors: | Ponnuraj, K, Rowland, S, Nessi, C, Setlow, P, Jedrzejas, M.J. | | Deposit date: | 2000-05-03 | | Release date: | 2001-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a novel germination protease from spores of Bacillus megaterium: structural arrangement and zymogen activation.
J.Mol.Biol., 300, 2000
|
|
1C82
 
 | |
1RTK
 
 | | Crystal Structure Analysis of the Bb segment of Factor B complexed with 4-guanidinobenzoic acid | | Descriptor: | 4-carbamimidamidobenzoic acid, Complement factor B Bb fragment, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1RRK
 
 | | Crystal Structure Analysis of the Bb segment of Factor B | | Descriptor: | COBALT (II) ION, Complement factor B, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-08 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
1RS0
 
 | | Crystal Structure Analysis of the Bb segment of Factor B complexed with Di-isopropyl-phosphate (DIP) | | Descriptor: | Complement factor B, DIISOPROPYL PHOSPHONATE, IODIDE ION, ... | | Authors: | Ponnuraj, K, Xu, Y, Macon, K, Moore, D, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 2003-12-09 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase.
Mol.Cell, 14, 2004
|
|
2OKM
 
 | |
1R19
 
 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (Apo structure) | | Descriptor: | fibrinogen-binding protein SdrG | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
1R17
 
 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (adhesin-ligand complex) | | Descriptor: | CALCIUM ION, fibrinogen-binding protein SdrG, fibrinopeptide B | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
4RMB
 
 | |
3LA4
 
 | |
4MR0
 
 | | Crystal structure of PfbA, a surface adhesin of Streptococcus pneumoniae | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ponnuraj, K, Beulin, D.S.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of PfbA, a surface adhesin of Streptococcus pneumoniae, provides hints into its interaction with fibronectin
Int.J.Biol.Macromol., 64C, 2013
|
|
5FCE
 
 | |
6IEP
 
 | | GAPDH of Streptococcus agalactiae | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Ponnuraj, K, Nagarajan, R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-01-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GAPDH of Streptococcus agalactiae and characterization of its interaction with extracellular matrix molecules
Microb. Pathog., 127, 2018
|
|
3V10
 
 | | Crystal structure of the collagen binding domain of Erysipelothrix rhusiopathiae surface protein RspB | | Descriptor: | Rhusiopathiae surface protein B | | Authors: | Ponnuraj, K, Swarmistha devi, A, Ogawa, Y, Shimoji, Y, Subramainan, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-10-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Collagen adhesin-nanoparticle interaction impairs adhesin's ligand binding mechanism
Biochim.Biophys.Acta, 1820, 2012
|
|
3HJT
 
 | |
2Z1P
 
 | |
2RAL
 
 | | Crystal Structure Analysis of double cysteine mutant of S.epidermidis adhesin SdrG: Evidence for the Dock,Lock and Latch ligand binding mechanism | | Descriptor: | Serine-aspartate repeat-containing protein G | | Authors: | Ponnuraj, K, Sthanam, N, Bowden, M.G, Hook, M. | | Deposit date: | 2007-09-17 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the "dock, lock, and latch" ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG.
J.Biol.Chem., 283, 2008
|
|
3L30
 
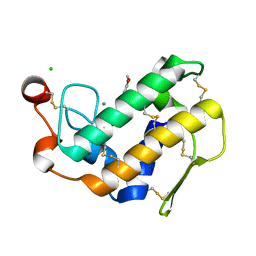 | | Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine | | Descriptor: | 4,14-dihydro-8H-[1,3]dioxolo[4,5-g]isoquino[3,2-a]isoquinoline-9,10-diol, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Naveen, C, Prasanth, G.K, Abhilash, J, Pradeep, M, Ponnuraj, K, Sadasivan, C, Haridas, M. | | Deposit date: | 2009-12-16 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of porcine pancreatic phospholipase A2 complexed with dihydroxyberberine
To be Published
|
|
4G7E
 
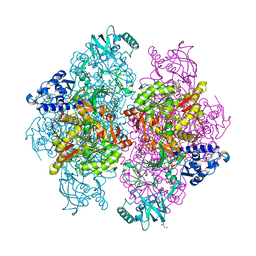 | |
4GOA
 
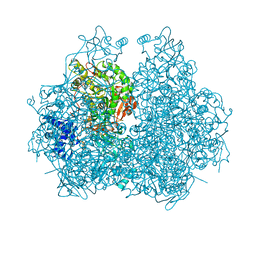 | |
3FRP
 
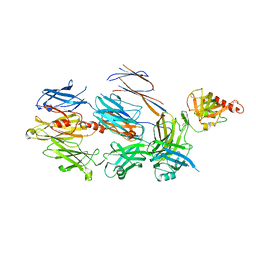 | | Crystal Structure of Cobra Venom Factor, a Co-factor for C3- and C5 convertase CVFBb | | Descriptor: | CALCIUM ION, Cobra venom factor alpha chain, Cobra venom factor beta chain, ... | | Authors: | Krishnan, V, Narayana, S.V.L. | | Deposit date: | 2009-01-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | The Crystal Structure of Cobra Venom Factor, a Cofactor for C3- and C5-Convertase CVFBb.
Structure, 17, 2009
|
|
2YX4
 
 | | Crystal Structure of T134A of ST1022 from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
2YX7
 
 | | Crystals structure of T132A mutant of St1022 from sulfolobus tokodaii 7 | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-24 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
2PMH
 
 | | Crystal structure of Thr132Ala of ST1022 from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION, ... | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-22 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
2PN6
 
 | | Crystal Structure of S32A of ST1022-Gln complex from Sulfolobus tokodaii | | Descriptor: | 150aa long hypothetical transcriptional regulator, GLUTAMINE, MAGNESIUM ION | | Authors: | Kumarevel, T.S, Karthe, P, Nakano, N, Shinkai, A, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structure of glutamine receptor protein from Sulfolobus tokodaii strain 7 in complex with its effector L-glutamine: implications of effector binding in molecular association and DNA binding
Nucleic Acids Res., 36, 2008
|
|
