6XT1
 
 | |
6XSX
 
 | |
6XJ9
 
 | | Structure of PfGH50B | | Descriptor: | 1,2-ETHANEDIOL, Agarase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2020-06-23 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of PfGH50B, an agarase from the marine bacterium Pseudoalteromonas fuliginea PS47
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6XSZ
 
 | |
2LTJ
 
 | | Conformational analysis of StrH, the surface-attached exo- beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae | | Descriptor: | Beta-N-acetylhexosaminidase | | Authors: | Pluvinage, B, Chitayat, S, Ficko-Blean, E, Abbott, D, Kunjachen, J, Grondin, J, Spencer, H, Smith, S, Boraston, A. | | Deposit date: | 2012-05-28 | | Release date: | 2012-11-28 | | Last modified: | 2013-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational analysis of StrH, the surface-attached exo-beta-D-N-acetylglucosaminidase from Streptococcus pneumoniae.
J.Mol.Biol., 425, 2013
|
|
5T99
 
 | |
5TA5
 
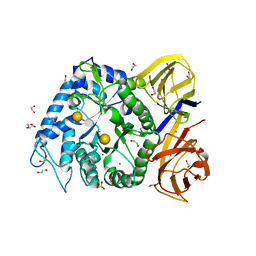 | | Crystal structure of BuGH86wt in complex with neoagarooctaose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5T9X
 
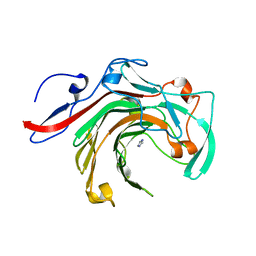 | | Crystal structure of BuGH16Bwt | | Descriptor: | Glycoside Hydrolase, IMIDAZOLE, SODIUM ION | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5UPI
 
 | |
5UPM
 
 | |
5V1W
 
 | | Crystal structure of BhGH81 in complex with laminaro-biose | | Descriptor: | 1,2-ETHANEDIOL, Glycoside Hydrolase, PHOSPHATE ION, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The quaternary structure of beta-1,3-glucan contributes to its recognition and hydrolysis by a multimodular family 81 glycoside hydrolase
Structure, 2017
|
|
5UPO
 
 | |
5UPN
 
 | |
7JNB
 
 | |
7JS4
 
 | |
7JND
 
 | |
7JNF
 
 | |
7JRM
 
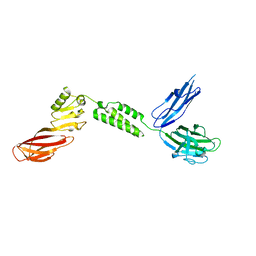 | |
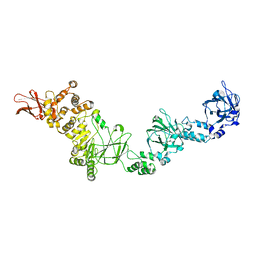
7JFS
 
 | |
7JRL
 
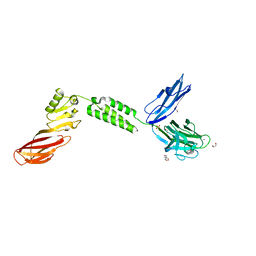 | | The structure of CBM51-2 in complex with GlcNAc and INT domains from Clostridium perfringens ZmpB | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2020-08-12 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecturally complex O -glycopeptidases are customized for mucin recognition and hydrolysis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5T98
 
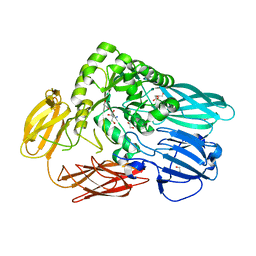 | | Crystal structure of BuGH2Awt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA1
 
 | | Crystal structure of BuGH86wt | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TA9
 
 | |
5T9G
 
 | |
5TA7
 
 | | Crystal structure of BuGH117Bwt | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoside Hydrolase, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
