2MQ9
 
 | |
2MQ6
 
 | |
2LGT
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for QFM(Y)F | | Descriptor: | Eukaryotic peptide chain release factor subunit 1 | | Authors: | Wong, L.E, Li, Y, Pillay, S, Pervushin, K. | | Deposit date: | 2011-08-02 | | Release date: | 2012-03-14 | | Method: | SOLUTION NMR | | Cite: | Selectivity of stop codon recognition in translation termination is modulated by multiple conformations of GTS loop in eRF1
Nucleic Acids Res., 2012
|
|
5XJK
 
 | |
2Q6Q
 
 | | Crystal structure of Spc42p, a critical component of spindle pole body in budding yeast | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Spindle pole body component SPC42 | | Authors: | Malashkevich, V.N, Newman, J.R, Kim, P.S. | | Deposit date: | 2007-06-05 | | Release date: | 2008-05-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Analysis of coiled-coil interactions between core proteins of the spindle pole body.
Biochemistry, 47, 2008
|
|
7RUU
 
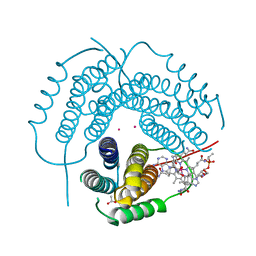 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, ACETATE ION, COBALAMIN, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUT
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase R190C bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Corrinoid adenosyltransferase, GLYCEROL, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
7RUV
 
 | | Structure of Human ATP:Cobalamin Adenosyltransferase E193K bound to adenosylcobalamin | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, Corrinoid adenosyltransferase, ... | | Authors: | Mascarenhas, R, Gouda, H, Koutmos, M, Banerjee, R. | | Deposit date: | 2021-08-18 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Patient mutations in human ATP:cob(I)alamin adenosyltransferase differentially affect its catalytic versus chaperone functions.
J.Biol.Chem., 297, 2021
|
|
