7KEZ
 
 | |
7KF0
 
 | |
7KF1
 
 | |
5I1V
 
 | |
5I1W
 
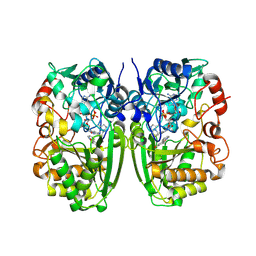 | | Crystal structure of CrmK, a flavoenzyme involved in the shunt product recycling mechanism in caerulomycin biosynthesis | | Descriptor: | 4-hydroxy[2,2'-bipyridine]-6-carbaldehyde, 6-(hydroxymethyl)[2,2'-bipyridin]-4-ol, CrmK, ... | | Authors: | Picard, M.-E, Barma, J, Shi, R. | | Deposit date: | 2016-02-07 | | Release date: | 2017-02-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural insights into flavoenzyme CrmK reveals a shunt product recycling mechanism in caerulomycin biosynthesis
to be published
|
|
6B02
 
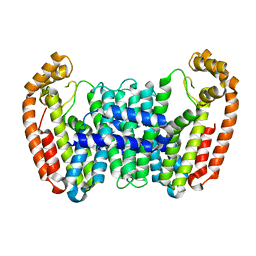 | |
6B07
 
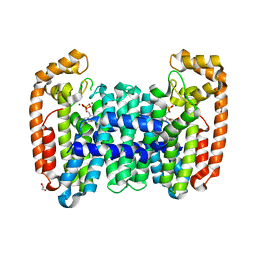 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [1-phosphono-2-(1-propylpyridin-2-yl)ethyl]phosphonic acid (inhibitor 1d) | | Descriptor: | 1,2-ETHANEDIOL, Farnesyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
6B06
 
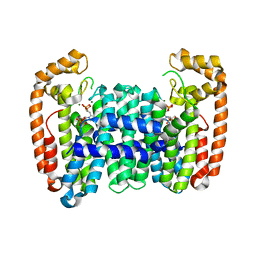 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with IPP and [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Farnesyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
6B04
 
 | | Crystal structure of CfFPPS2, a lepidopteran type-II farnesyl diphosphate synthase, complexed with [2-(1-methylpyridin-2-yl)-1-phosphono-ethyl]phosphonic acid (inhibitor 1b) | | Descriptor: | 1,2-ETHANEDIOL, Farnesyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Picard, M.-E, Cusson, M, Shi, R. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural characterization of a lepidopteran type-II farnesyl diphosphate synthase from the spruce budworm, Choristoneura fumiferana: Implications for inhibitor design.
Insect Biochem. Mol. Biol., 92, 2017
|
|
6MXS
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98F,HC-G99M] | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | Authors: | Shi, R, Picard, M.-E, Manenda, M.S. | | Deposit date: | 2018-10-31 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6MY4
 
 | | Crystal structure of the dimeric bH1-Fab variant [HC-Y33W,HC-D98M,HC-G99M,LC-S30bR] | | Descriptor: | 1,2-ETHANEDIOL, anti-VEGF-A Fab fragment bH1 heavy chain, anti-VEGF-A Fab fragment bH1 light chain | | Authors: | Shi, R, Picard, M.-E, Manenda, M. | | Deposit date: | 2018-11-01 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Binding symmetry and surface flexibility mediate antibody self-association.
Mabs, 11, 2019
|
|
6U0P
 
 | | Crystal structure of PieE, the flavin-dependent monooxygenase involved in the biosynthesis of piericidin A1 | | Descriptor: | 2,4-dichlorophenol 6-monooxygenase, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Shi, R, Manenda, M, Picard, M.-E. | | Deposit date: | 2019-08-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural analyses of the Group A flavin-dependent monooxygenase PieE reveal a sliding FAD cofactor conformation bridging OUT and IN conformations.
J.Biol.Chem., 295, 2020
|
|
8D8W
 
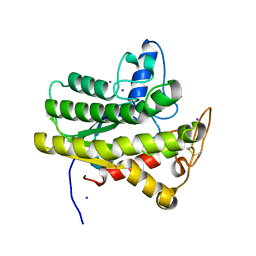 | | Crystal structure of ChoE with Ser38 adopting alternative conformations | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ChoE, IODIDE ION | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8Y
 
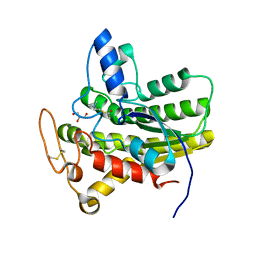 | | Crystal structure of ChoE N147A mutant in complex with acetylthiocholine | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ACETYLTHIOCHOLINE, CHLORIDE ION, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D91
 
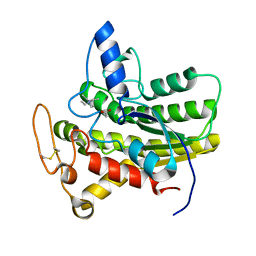 | | Crystal structure of ChoE in complex with acetate and tetraethylammonium (TEA) | | Descriptor: | ACETATE ION, ChoE, TETRAETHYLAMMONIUM ION | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8X
 
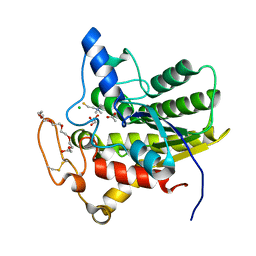 | | Crystal structure of ChoE in complex with acetate and thiocholine (crystal form 2) | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D8Z
 
 | | Crystal structure of ChoE N147A mutant in complex with thiocholine and chloride | | Descriptor: | 2-(TRIMETHYLAMMONIUM)ETHYL THIOL, CHLORIDE ION, ChoE, ... | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
8D90
 
 | | Crystal structure of ChoE N147A mutant in complex with bromide ions | | Descriptor: | BROMIDE ION, ChoE, GLYCEROL | | Authors: | Pham, V.D, Shi, R. | | Deposit date: | 2022-06-09 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of bacterial acetylcholinesterase ChoE provide insights into the plasticity of catalytic Ser in regulating the active site geometry and the functional state of the SGNH hydrolases
To be published
|
|
