3IGC
 
 | | Smallpox virus topoisomerase-DNA transition state | | Descriptor: | 5'-D(*AP*TP*TP*CP*C)-3', 5'-D(*CP*GP*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*GP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', ... | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2009-07-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic.
Structure, 18, 2010
|
|
2H7F
 
 | | Structure of variola topoisomerase covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
2H7G
 
 | | Structure of variola topoisomerase non-covalently bound to DNA | | Descriptor: | 5'-D(*TP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*TP*TP*GP*TP*CP*GP*CP*CP*CP*TP*TP*A)-3', DNA topoisomerase 1 | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2006-06-02 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for specificity in the poxvirus topoisomerase.
Mol.Cell, 23, 2006
|
|
1MW8
 
 | |
1MW9
 
 | |
5JMC
 
 | | Receptor binding domain of Botulinum neurotoxin A in complex with rat SV2C | | Descriptor: | Botulinum neurotoxin type A, Synaptic vesicle glycoprotein 2C | | Authors: | Yao, G, Zhang, S, Mahrhold, S, Lam, K, Stern, D, Bagramyan, K, Perry, K, Kalkum, M, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5JLV
 
 | | Receptor binding domain of Botulinum neurotoxin A in complex with human glycosylated SV2C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Yao, G, Zhang, S, Mahrhold, S, Lam, K, Stern, D, Bagramyan, K, Perry, K, Kalkum, M, Rummel, A, Dong, M, Jin, R. | | Deposit date: | 2016-04-27 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-linked glycosylation of SV2 is required for binding and uptake of botulinum neurotoxin A.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8VRS
 
 | | Mucin 16 peptide fused to MBP in complex with 4H11-scFv antibody | | Descriptor: | 4H11 scFv chain, Maltose/maltodextrin-binding periplasmic protein,Mucin-16 | | Authors: | Lee, K, Perry, K, Yeku, O.O, Spriggs, D.R. | | Deposit date: | 2024-01-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural basis for antibody recognition of the proximal MUC16 ectodomain.
J Ovarian Res, 17, 2024
|
|
8VRR
 
 | | 4H11-scFv antibody | | Descriptor: | 4H11 scFv chain | | Authors: | Lee, K, Perry, K, Yeku, O.O, Spriggs, D.R. | | Deposit date: | 2024-01-22 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for antibody recognition of the proximal MUC16 ectodomain.
J Ovarian Res, 17, 2024
|
|
7C0Q
 
 | | a Legionella pneumophila effector Lpg2505 | | Descriptor: | GLYCEROL, effector Lpg2505 | | Authors: | Chen, T.T, Han, A.D, Luo, Z.Q, McCloskey, A, Perri, K. | | Deposit date: | 2020-05-01 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The metaeffector MesI regulates the activity of the Legionella effector SidI through direct protein-protein interactions.
Microbes Infect., 23, 2021
|
|
8UIV
 
 | | H47Q NicC with bound FAD | | Descriptor: | 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hicks, K.A, Perry, K. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.511 Å) | | Cite: | Ligand bound structure of a 6-hydroxynicotinic acid 3-monooxygenase provides mechanistic insights.
Arch.Biochem.Biophys., 752, 2024
|
|
8UIQ
 
 | | H47Q NicC with 2-mercaptopyridine ligand | | Descriptor: | 2-PYRIDINETHIOL, 6-hydroxynicotinate 3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Hicks, K.A, Perry, K, Turlington, Z.R, Vaz Ferreira de Macedo, S. | | Deposit date: | 2023-10-10 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Ligand bound structure of a 6-hydroxynicotinic acid 3-monooxygenase provides mechanistic insights.
Arch.Biochem.Biophys., 752, 2024
|
|
6EDI
 
 | |
4KIS
 
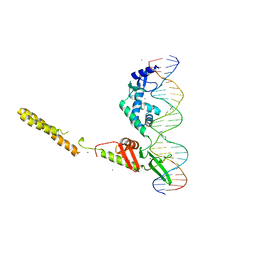 | | Crystal Structure of a LSR-DNA Complex | | Descriptor: | CALCIUM ION, DNA (26-MER), Putative integrase [Bacteriophage A118], ... | | Authors: | Rutherford, K, Yuan, P, Perry, K, Van Duyne, G.D. | | Deposit date: | 2013-05-02 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Attachment site recognition and regulation of directionality by the serine integrases.
Nucleic Acids Res., 41, 2013
|
|
4LO3
 
 | | HA17-HA33-LacNac | | Descriptor: | HA-17, HA-33, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
4LO0
 
 | | Apo HA17-HA33 | | Descriptor: | HA-17, HA-33 | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
5E6F
 
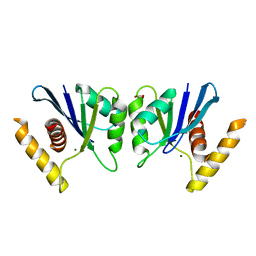 | | Canarypox virus resolvase | | Descriptor: | CNPV261 Holliday junction resolvase protein, D(-)-TARTARIC ACID, MAGNESIUM ION | | Authors: | Li, H, Hwang, Y, Perry, K, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Metal Binding Properties of a Poxvirus Resolvase.
J.Biol.Chem., 291, 2016
|
|
4GLI
 
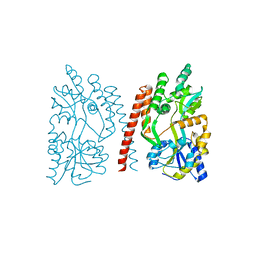 | | Crystal Structure of Human SMN YG-Dimer | | Descriptor: | Maltose-binding periplasmic protein, Survival motor neuron protein chimera | | Authors: | Martin, R.S, Perry, K, Van Duyne, G.D. | | Deposit date: | 2012-08-14 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The survival motor neuron protein forms soluble glycine zipper oligomers.
Structure, 20, 2012
|
|
5FCW
 
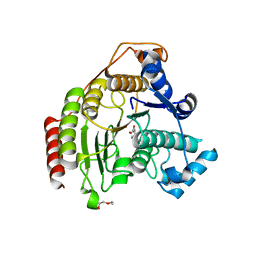 | | HDAC8 Complexed with a Hydroxamic Acid | | Descriptor: | 4-naphthalen-1-yl-~{N}-oxidanyl-benzamide, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Cole, K.E, Perry, K. | | Deposit date: | 2015-12-15 | | Release date: | 2016-10-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structure of 'linkerless' hydroxamic acid inhibitor-HDAC8 complex confirms the formation of an isoform-specific subpocket.
J.Struct.Biol., 195, 2016
|
|
4E1H
 
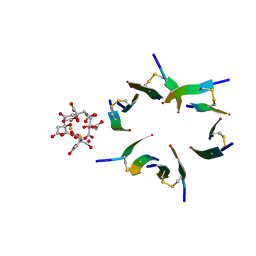 | | Fragment of human prion protein | | Descriptor: | CITRIC ACID, FE (III) ION, Major prion protein | | Authors: | Apostol, M.I, Perry, K, Surewicz, W.K. | | Deposit date: | 2012-03-06 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of a human prion protein fragment reveals a motif for oligomer formation.
J.Am.Chem.Soc., 135, 2013
|
|
3IX1
 
 | | Periplasmic N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding protein from Bacillus halodurans | | Descriptor: | N-[(4-amino-2-methylpyrimidin-5-yl)methyl]formamide, N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding protein | | Authors: | Bale, S, Rajashankar, K.R, Perry, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-09-03 | | Release date: | 2010-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | HMP Binding Protein ThiY and HMP-P Synthase THI5 Are Structural Homologues.
Biochemistry, 49, 2010
|
|
4LO2
 
 | | HA17-HA33-Lac | | Descriptor: | HA-17, HA-33, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
4LO4
 
 | | Apo HA-70 | | Descriptor: | CHLORIDE ION, HA-70 | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.871 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
4LO1
 
 | | HA17-HA33-Gal | | Descriptor: | HA-17, HA-33, beta-D-galactopyranose | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
4LO5
 
 | | HA70-alpha2,3-SiaLC | | Descriptor: | CHLORIDE ION, HA-70, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
