5UPW
 
 | |
5UP4
 
 | |
5UOT
 
 | |
3J3Y
 
 | |
3J3Q
 
 | |
6X63
 
 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-05-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6T64
 
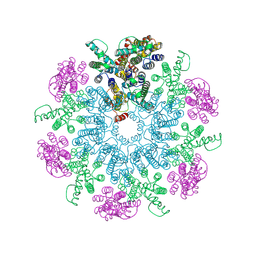 | | A model of the EIAV CA-SP hexamer (C6) from Gag-deltaMA spheres assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T63
 
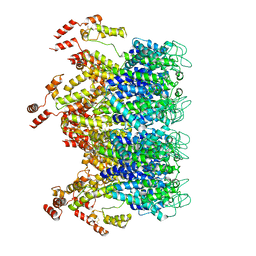 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH6 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
6T61
 
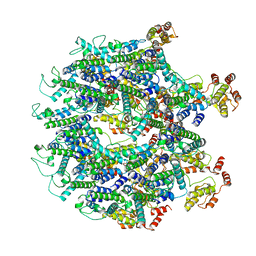 | | A model of the EIAV CA-SP hexamer (C2) from Gag-deltaMA tubes assembled at pH8 | | Descriptor: | Gag polyprotein | | Authors: | Dick, R.A, Xu, C, Morado, D.R, Kravchuk, V, Ricana, C.L, Lyddon, T.D, Broad, A.M, Feathers, J.R, Johnson, M.C, Vogt, V.M, Perilla, J.R, Briggs, J.A.G, Schur, F.K.M. | | Deposit date: | 2019-10-17 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly.
Plos Pathog., 16, 2020
|
|
7U8K
 
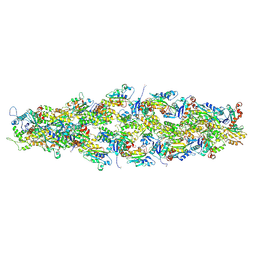 | | Magic Angle Spinning NMR Structure of Human Cofilin-2 Assembled on Actin Filaments | | Descriptor: | Actin, alpha skeletal muscle, Cofilin-2 | | Authors: | Kraus, J, Russell, R, Kudryashova, E, Xu, C, Katyal, N, Kudryashov, D, Perilla, J.R, Polenova, T. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | SOLID-STATE NMR | | Cite: | Magic angle spinning NMR structure of human cofilin-2 assembled on actin filaments reveals isoform-specific conformation and binding mode.
Nat Commun, 13, 2022
|
|
5FJB
 
 | | Cyclophilin A Stabilize HIV-1 Capsid through a Novel Non- canonical Binding Site | | Descriptor: | GAG POLYPROTEIN, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Liu, C, Perilla, J.R, Ning, J, Lu, M, Hou, G, Ramalhu, R, Bedwell, G.J, Ahn, J, Shi, J, Gronenborn, A.M, Prevelige Jr, P.E, Rousso, I, Aiken, C, Polenova, T, Schulten, K, Zhang, P. | | Deposit date: | 2015-10-07 | | Release date: | 2016-03-16 | | Last modified: | 2017-08-23 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Cyclophilin a Stabilizes the HIV-1 Capsid Through a Novel Non-Canonical Binding Site.
Nat.Commun., 7, 2016
|
|
7R7Q
 
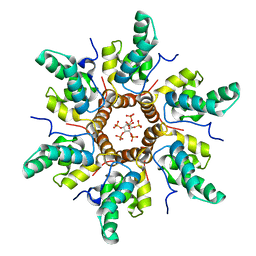 | | Immature HIV-1 CACTD-SP1 lattice with Inositol hexakisphosphate (IP6) | | Descriptor: | Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-22 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
7R7P
 
 | | Immature HIV-1 CACTD-SP1 lattice with Bevirimat (BVM) and Inositol hexakisphosphate (IP6) | | Descriptor: | 3alpha-[(3-carboxy-3-methylbutanoyl)oxy]-8alpha,9beta,10alpha,13alpha,17alpha,19beta-lup-20(29)-en-28-oic acid, Gag polyprotein, INOSITOL HEXAKISPHOSPHATE | | Authors: | Sarkar, S, Zadrozny, K.K, Zadorozhnyi, R, Russell, R.W, Quinn, C.M, Kleinpeter, A, Ablan, S, Meshkin, H, Perilla, J.R, Ganser-Pornillos, B.K, Pornillos, O, Freed, E.O, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-06-25 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-22 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of HIV-1 maturation inhibitor binding and activity.
Nat Commun, 14, 2023
|
|
8C9M
 
 | | HERV-K Gag immature lattice | | Descriptor: | Gag protein | | Authors: | Krebs, A.-S, Liu, H.-F, Zhou, Y, Rey, J.S, Levintov, L, Perilla, J.R, Bartesaghi, A, Zhang, P. | | Deposit date: | 2023-01-23 | | Release date: | 2023-02-01 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and conservation of an immature human endogenous retrovirus.
Biorxiv, 2023
|
|
6WAP
 
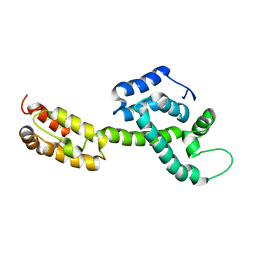 | | Atomic-Resolution Structure of HIV-1 Capsid Tubes by Magic Angle Spinning NMR | | Descriptor: | HIV-1 capsid protein | | Authors: | Lu, M, Russell, R.W, Bryer, A, Quinn, C.M, Hou, G, Zhang, H, Schwieters, C.D, Perilla, J.R, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2020-03-25 | | Release date: | 2020-09-02 | | Last modified: | 2023-06-14 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of HIV-1 capsid tubes by magic-angle spinning NMR.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3JA6
 
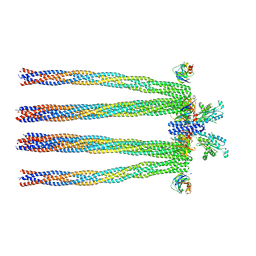 | | Cryo-electron Tomography and All-atom Molecular Dynamics Simulations Reveal a Novel Kinase Conformational Switch in Bacterial Chemotaxis Signaling | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein 2 | | Authors: | Cassidy, C.K, Himes, B.A, Alvarez, F.J, Ma, J, Zhao, G, Perilla, J.R, Schulten, K, Zhang, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Elife, 4, 2015
|
|
3J4F
 
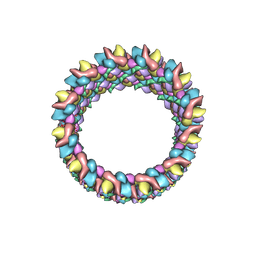 | | Structure of HIV-1 capsid protein by cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Meng, X, Schulten, K, Zhang, P. | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
6OMT
 
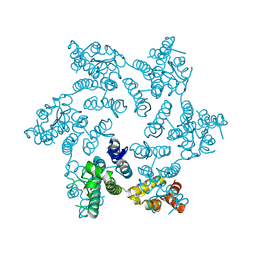 | | HIV-1 capsid hexamer R18D mutant | | Descriptor: | Capsid protein | | Authors: | Huang, P, Summers, B.J, Xiong, Y. | | Deposit date: | 2019-04-19 | | Release date: | 2019-08-21 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | FEZ1 Is Recruited to a Conserved Cofactor Site on Capsid to Promote HIV-1 Trafficking.
Cell Rep, 28, 2019
|
|
7M0G
 
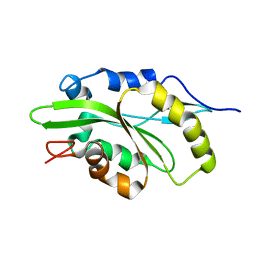 | |
6E7E
 
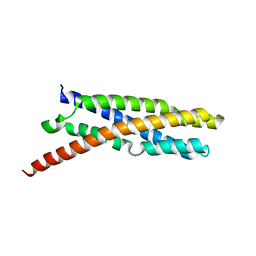 | |
6E6A
 
 | | Triclinic crystal form of IncA G144A point mutant | | Descriptor: | Inclusion membrane protein A, SODIUM ION | | Authors: | Cingolani, G, Paumet, F. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the homotypic fusion of chlamydial inclusions by the SNARE-like protein IncA.
Nat Commun, 10, 2019
|
|
8A1Q
 
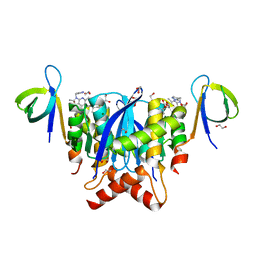 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor STP0404 (Pirmitegravir) | | Descriptor: | (2S)-tert-butoxy{4-(4-chlorophenyl)-2,3,6-trimethyl-1-[(1-methyl-1H-pyrazol-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridin-5-yl}acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
8A1P
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BI-D | | Descriptor: | (2S)-tert-butoxy[4-(3,4-dihydro-2H-chromen-6-yl)-2-methylquinolin-3-yl]ethanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, M.R, Pye, V.E, Cook, N.J, Cherepanov, P. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Drug-Induced Interface That Drives HIV-1 Integrase Hypermultimerization and Loss of Function.
Mbio, 14, 2023
|
|
7N9H
 
 | |
