1AOK
 
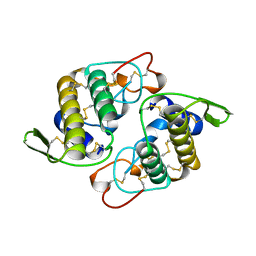 | | VIPOXIN COMPLEX | | Descriptor: | ACETATE ION, VIPOXIN COMPLEX | | Authors: | Perbandt, M, Wilson, J.C, Eschenburg, S, Betzel, C. | | Deposit date: | 1997-07-07 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of vipoxin at 2.0 A: an example of regulation of a toxic function generated by molecular evolution.
FEBS Lett., 412, 1997
|
|
1LNL
 
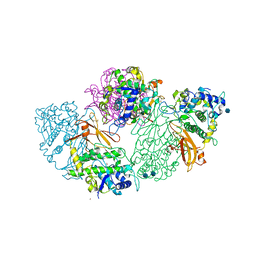 | | Structure of deoxygenated hemocyanin from Rapana thomasiana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, hemocyanin | | Authors: | Perbandt, M, Guthoehrlein, E.W, Rypniewski, W, Idakieva, K, Stoeva, S, Voelter, W, Genov, N, Betzel, C. | | Deposit date: | 2002-05-03 | | Release date: | 2003-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity
Biochemistry, 42, 2003
|
|
1TU8
 
 | |
2G5L
 
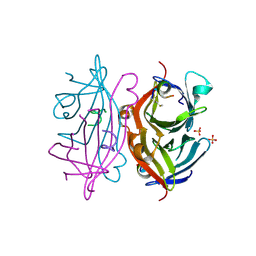 | | Streptavidin in complex with Nanotag | | Descriptor: | (FME)(ASP)(VAL)(GLU)(ALA)(TRP)(LEU), GLYCEROL, SULFATE ION, ... | | Authors: | Perbandt, M, Bruns, O, Vallazza, M, Lamla, T, Betzel, C, Erdmann, V.A. | | Deposit date: | 2006-02-23 | | Release date: | 2007-02-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High resolution structure of Streptavidin in complex with a novel high affinity peptide tag mimicking the biotin binding motif
To be Published
|
|
4ZXG
 
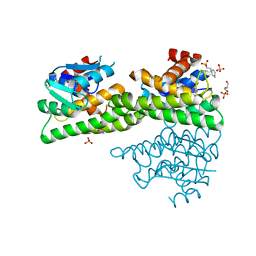 | | Ligandin binding site of PfGST | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Perbandt, M, Eberle, R, Betzel, C. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution structures of Plasmodium falciparum GST complexes provide novel insights into the dimer-tetramer transition and a novel ligand-binding site.
J.Struct.Biol., 191, 2015
|
|
361D
 
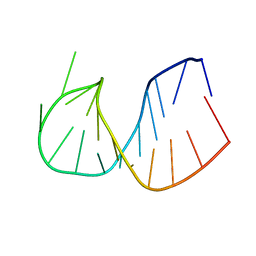 | | CRYSTAL STRUCTURE OF DOMAIN E OF THERMUS FLAVUS 5S RRNA: A HELICAL RNA-STRUCTURE INCLUDING A TETRALOOP | | Descriptor: | RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*GP*GP*CP*GP*AP*CP*CP*GP*CP*C P*UP*GP*G)-3') | | Authors: | Perbandt, M, Nolte, A, Lorenz, S, Erdmann, V.A, Betzel, C. | | Deposit date: | 1997-11-10 | | Release date: | 1998-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of domain E of Thermus flavus 5S rRNA: a helical RNA structure including a hairpin loop.
FEBS Lett., 429, 1998
|
|
3FRC
 
 | |
3FR9
 
 | |
3FR6
 
 | |
3FR3
 
 | |
439D
 
 | | 5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3', 5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3' | | Descriptor: | BARIUM ION, RNA (5'-R(*CP*CP*GP*CP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*UP*GP*GP*GP*CP*GP*G)-3') | | Authors: | Perbandt, M, Lorenz, S, Vallazza, M, Erdmann, V.A, Betzel, C. | | Deposit date: | 1999-01-05 | | Release date: | 2001-09-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of an RNA duplex with an unusual G.C pair in wobble-like conformation at 1.6 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2HNL
 
 | | Structure of the prostaglandin D synthase from the parasitic nematode Onchocerca volvulus | | Descriptor: | GLUTATHIONE, Glutathione S-transferase 1 | | Authors: | Perbandt, M, Hoppner, J, Betzel, C, Liebau, E. | | Deposit date: | 2006-07-13 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the extracellular glutathione S-transferase OvGST1 from the human pathogenic parasite Onchocerca volvulus.
J.Mol.Biol., 377, 2008
|
|
1OQS
 
 | | Crystal Structure of RV4/RV7 Complex | | Descriptor: | Phospholipase A2 RV-4, Phospholipase A2 RV-7 | | Authors: | Perbandt, M, Betzel, C. | | Deposit date: | 2003-03-11 | | Release date: | 2003-09-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the heterodimeric neurotoxic complex viperotoxin F (RV-4/RV-7) from the venom of Vipera russelli formosensis at 1.9 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1PA3
 
 | |
1Q4J
 
 | |
1TU7
 
 | |
7Q11
 
 | | Crystal structure of CTX-M-14 in complex with Ixazomib | | Descriptor: | Beta-lactamase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
7Q0Y
 
 | | Crystal structure of CTX-M-14 in complex with Bortezomib | | Descriptor: | ACETATE ION, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
7Q0Z
 
 | | Crystal structure of CTX-M-14 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Werner, N, Perbandt, M, Hinrichs, W, Prester, A, Rohde, H, Aepfelbacher, M, Betzel, C. | | Deposit date: | 2021-10-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural basis to repurpose boron-based proteasome inhibitors Bortezomib and Ixazomib as beta-lactamase inhibitors.
Sci Rep, 12, 2022
|
|
3R3L
 
 | | Structure of NP protein from Lassa AV strain | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, ZINC ION | | Authors: | Perbandt, M, Brunotte, L, Gunther, S, Betzel, C. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Structure of the Lassa virus nucleoprotein revealed by X-ray crystallography, small-angle X-ray scattering, and electron microscopy.
J.Biol.Chem., 286, 2011
|
|
1QL0
 
 | |
4UQF
 
 | | CRYSTAL STRUCTURE OF LISTERIA MONOCYTOGENES GTP CYCLOHYDROLASE I | | Descriptor: | GTP cyclohydrolase 1 | | Authors: | Schuessler, S, Perbandt, M, Fischer, M, Graewert, T. | | Deposit date: | 2014-06-23 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of GTP cyclohydrolase I from Listeria monocytogenes, a potential anti-infective drug target.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1CE7
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
6RFU
 
 | | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of ATP and GMP as genuine co-factors | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Inosine-5'-monophosphate dehydrogenase | | Authors: | Nass, K, Redecke, L, Perbandt, M, Yefanov, O, Gabdulkhakov, A, Duszenko, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In cellulo crystallization of Trypanosoma brucei IMP dehydrogenase enables the identification of genuine co-factors.
Nat Commun, 11, 2020
|
|
4OW2
 
 | | YopM from Yersinia enterocolitica WA-314 | | Descriptor: | Yop effector YopM | | Authors: | Rumm, A, Perbandt, M, Aepfelbacher, M. | | Deposit date: | 2014-01-30 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Immunosuppressive Yersinia Effector YopM Binds DEAD Box Helicase DDX3 to Control Ribosomal S6 Kinase in the Nucleus of Host Cells.
PLoS Pathog., 12, 2016
|
|
