2VB2
 
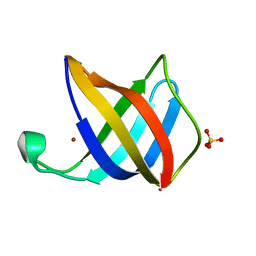 | | Crystal structure of Cu(I)CusF | | Descriptor: | CATION EFFLUX SYSTEM PROTEIN CUSF, COPPER (II) ION, SULFATE ION | | Authors: | Xue, Y, Davis, A.V, Balakrishnan, G, Stasser, J.P, Staehlin, B.M, Focia, P, Spiro, T.G, Penner-Hahn, J.E, O'Halloran, T.V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cu(I) Recognition Via Cation-Pi and Methionine Interactions in Cusf.
Nat.Chem.Biol., 4, 2008
|
|
2VB3
 
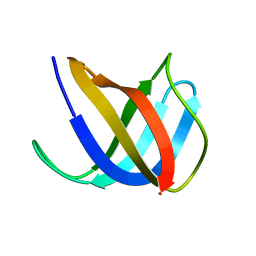 | | Crystal structure of Ag(I)CusF | | Descriptor: | CATION EFFLUX SYSTEM PROTEIN CUSF, SILVER ION | | Authors: | Xue, Y, Davis, A.V, Balakrishnan, G, Stasser, J.P, Staehlin, B.M, Focia, P, Spiro, T.G, Penner-Hahn, J.E, O'Halloran, T.V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Cu(I) Recognition Via Cation-Pi and Methionine Interactions in Cusf.
Nat.Chem.Biol., 4, 2008
|
|
1EYF
 
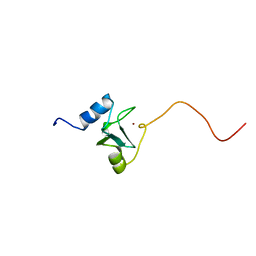 | | REFINED STRUCTURE OF THE DNA METHYL PHOSPHOTRIESTER REPAIR DOMAIN OF E. COLI ADA | | Descriptor: | ADA REGULATORY PROTEIN, ZINC ION | | Authors: | Lin, Y, Dotsch, V, Wintner, T, Peariso, K, Myers, L.C, Penner-Hahn, J.E, Verdine, G.L, Wagner, G. | | Deposit date: | 2000-05-06 | | Release date: | 2003-09-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the functional switch of the E. coli Ada protein
Biochemistry, 40, 2001
|
|
3K7R
 
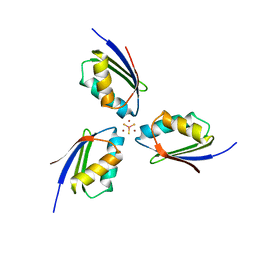 | | Crystal structure of [TM][CuAtx1]3 | | Descriptor: | COPPER (II) ION, D-MALATE, Metal homeostasis factor ATX1, ... | | Authors: | Xue, Y, Alvarez, H.M, Robinson, C.D, Mondragon, A, O'Halloran, T.V. | | Deposit date: | 2009-10-13 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Tetrathiomolybdate inhibits copper trafficking proteins through metal cluster formation.
Science, 327, 2010
|
|
6DS9
 
 | | Elongated version of a de novo designed three helix bundle structure (GRa3D) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THIOCYANATE ION, ... | | Authors: | Koebke, K.J, Ruckthong, L.R, Meagher, J.L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Clarifying the Copper Coordination Environment in a de Novo Designed Red Copper Protein.
Inorg Chem, 57, 2018
|
|
7N2Y
 
 | |
7N2Z
 
 | | Crystal Structure of a de Novo Three-stranded Coiled Coil Peptide Containing Trigonal Pyrmidal Pb(II) complexes in the dual Tris-thiolate Site | | Descriptor: | CHLORIDE ION, LEAD (II) ION, Pb(II)2-(GRAND CoilSerL16CL23C)3, ... | | Authors: | Ruckthong, L, Stuckey, J.A, Pecoraro, V.L. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Open Reading Frame 1 Protein of the Human Long Interspersed Nuclear Element 1 Retrotransposon Binds Multiple Equivalents of Lead.
J.Am.Chem.Soc., 143, 2021
|
|
5TIR
 
 | | Crystal Structure of Mn Superoxide Dismutase mutant M27V from Trichoderma reesei | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Mendoza, E.R, Brandao-Neto, J, Pereira, H.M, Ferreira Junior, J.R.S, Garratt, R.C. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of Mn Superoxide Dismutase mutant M27V from Trichoderma reesei
To Be Published
|
|
5FD1
 
 | |
1ZCP
 
 | |
2FD2
 
 | |
1GPX
 
 | | C85S GAPDX, NMR, 20 STRUCTURES | | Descriptor: | GALLIUM (III) ION, PUTIDAREDOXIN | | Authors: | Pochapsky, T.C, Kuti, M, Kazanis, S. | | Deposit date: | 1998-06-10 | | Release date: | 1999-01-27 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a gallium-substituted putidaredoxin mutant: GaPdx C85S.
J.Biomol.NMR, 12, 1998
|
|
1GPM
 
 | | ESCHERICHIA COLI GMP SYNTHETASE COMPLEXED WITH AMP AND PYROPHOSPHATE | | Descriptor: | ADENOSINE MONOPHOSPHATE, CITRIC ACID, GMP SYNTHETASE, ... | | Authors: | Tesmer, J.J.G. | | Deposit date: | 1995-04-04 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of GMP synthetase reveals a novel catalytic triad and is a structural paradigm for two enzyme families.
Nat.Struct.Biol., 3, 1996
|
|
1FDD
 
 | |
1FDB
 
 | |
1FD2
 
 | |
1FDA
 
 | |
