3BAN
 
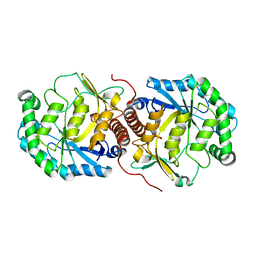 | | The crystal structure of mannonate dehydratase from Streptococcus suis serotype2 | | Descriptor: | D-mannonate dehydratase | | Authors: | Peng, H, Zhang, Q.M, Gao, F, Liu, Y.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2007-11-08 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of mannonate dehydratase from Streptococcus suis serotype2
To be Published
|
|
3FVM
 
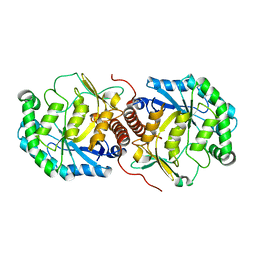 | | Crystal structure of Steptococcus suis mannonate dehydratase with metal Mn++ | | Descriptor: | MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q.J, Gao, F, Liu, Y, Gao, F.G. | | Deposit date: | 2009-01-16 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
3CXR
 
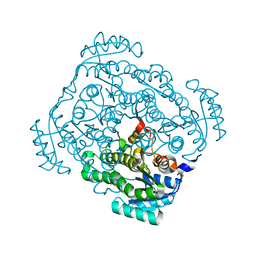 | | Crystal structure of gluconate 5-dehydrogase from streptococcus suis type 2 | | Descriptor: | Dehydrogenase with different specificities | | Authors: | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | Deposit date: | 2008-04-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into the catalytic mechanism of gluconate 5-dehydrogenase from Streptococcus suis: Crystal structures of the substrate-free and quaternary complex enzymes.
Protein Sci., 18, 2009
|
|
3O03
 
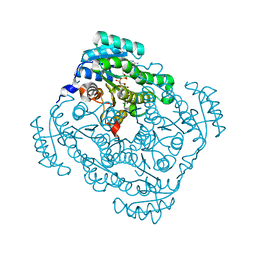 | | Quaternary complex structure of gluconate 5-dehydrogenase from streptococcus suis type 2 | | Descriptor: | CALCIUM ION, D-gluconic acid, Dehydrogenase with different specificities, ... | | Authors: | Peng, H, Gao, F, Zhang, Q, Liu, Y, Gao, G.F. | | Deposit date: | 2010-07-18 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight Into the Catalytic Mechanism of Gluconate 5-Dehydrogenase from Streptococcus Suis: Crystal Structures of the Substrate-Free and Quaternary Complex Enzymes.
Protein Sci., 18, 2009
|
|
3DBN
 
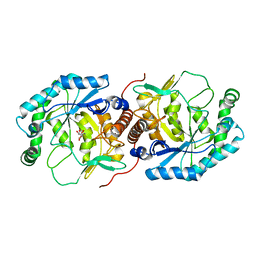 | | Crystal structure of the Streptoccocus suis serotype2 D-mannonate dehydratase in complex with its substrate | | Descriptor: | D-MANNONIC ACID, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q, Gao, F, Gao, G.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
2Q3A
 
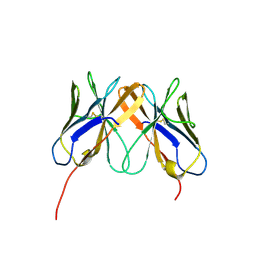 | | Crystal Structure of Rhesus Macaque CD8 Alpha-Alpha Homodimer | | Descriptor: | CD8 | | Authors: | Peng, H, Chen, Y, Zong, L, Gao, F, Liu, Y, Gao, G.F. | | Deposit date: | 2007-05-30 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Rhesus Macaque CD8 Homodimer Sheds the Lights on the Molecular Basis of Rhesus Macaque MHC class I with CD8 Complex
To be Published
|
|
6UJV
 
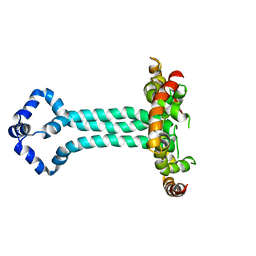 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
3GLR
 
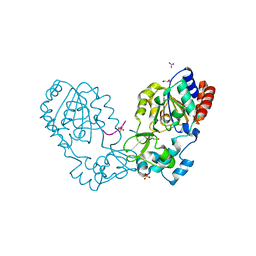 | | Crystal Structure of human SIRT3 with acetyl-lysine AceCS2 peptide | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, BICARBONATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3GLT
 
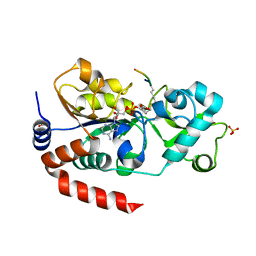 | | Crystal Structure of Human SIRT3 with ADPR bound to the AceCS2 peptide containing a thioacetyl lysine | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, CARBONATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3GLU
 
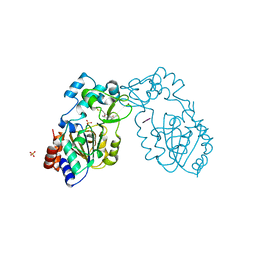 | | Crystal Structure of Human SIRT3 with AceCS2 peptide | | Descriptor: | Acetyl-coenzyme A synthetase 2-like, mitochondrial, NAD-dependent deacetylase sirtuin-3, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
3GLS
 
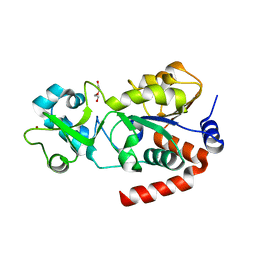 | | Crystal Structure of Human SIRT3 | | Descriptor: | NAD-dependent deacetylase sirtuin-3, mitochondrial, SULFATE ION, ... | | Authors: | Jin, L, Wei, W, Jiang, Y, Peng, H, Cai, J, Mao, C, Dai, H, Bemis, J.E, Jirousek, M.R, Milne, J.C, Westphal, C.H, Perni, R.B. | | Deposit date: | 2009-03-12 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes.
J.Biol.Chem., 284, 2009
|
|
6E8W
 
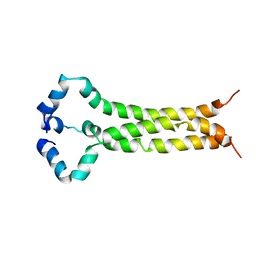 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4R5Y
 
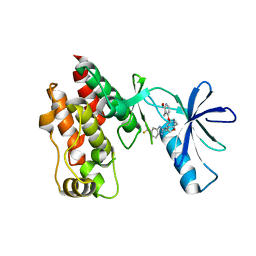 | | The complex structure of Braf V600E kinase domain with a novel Braf inhibitor | | Descriptor: | 5-({(1R,1aS,6bR)-1-[5-(trifluoromethyl)-1H-benzimidazol-2-yl]-1a,6b-dihydro-1H-cyclopropa[b][1]benzofuran-5-yl}oxy)-3,4-dihydro-1,8-naphthyridin-2(1H)-one, Serine/threonine-protein kinase B-raf | | Authors: | Feng, Y, Peng, H, Zhang, Y, Liu, Y, Wei, M. | | Deposit date: | 2014-08-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers.
Mol.Cancer Ther., 14, 2015
|
|
6UJU
 
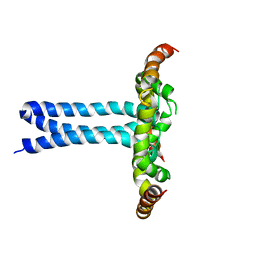 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
3BDK
 
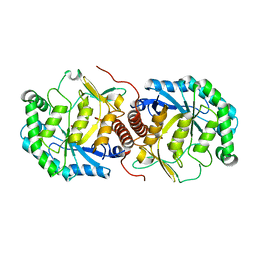 | | Crystal Structure of Streptococcus suis mannonate dehydratase complexed with substrate analogue | | Descriptor: | D-mannonate dehydratase, D-mannose, MANGANESE (II) ION | | Authors: | Gao, F, Zhang, Q.M, Peng, H, Liu, Y.W, Qi, J.X, Gao, G.F. | | Deposit date: | 2007-11-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Streptococcus suis mannonate dehydratase complexed with substrate analogue
To be Published
|
|
2OTP
 
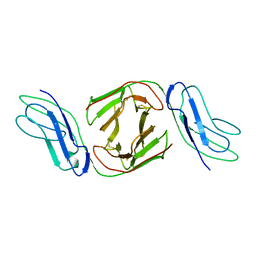 | | Crystal Structure of Immunoglobulin-Like Transcript 1 (ILT1/LIR7/LILRA2) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily A member 2 | | Authors: | Gao, F, Peng, H, Chen, Y, Liu, Y, Gao, G.F. | | Deposit date: | 2007-02-08 | | Release date: | 2008-02-19 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of 3D Domain Swapped Dimer of Immunoglobulin-Like Transcript 1 (ILT1/LIR7/LILRA2), Molecular Insight into Group 1 Activating Receptor Forming Unique MW Interaction Pattern
To be Published
|
|
3FAN
 
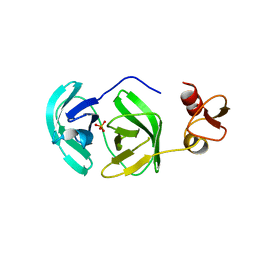 | | Crystal structure of chymotrypsin-like protease/proteinase (3CLSP/Nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV) | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
3FAO
 
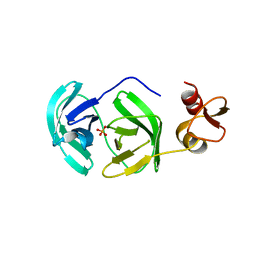 | | Crystal structure of S118A mutant 3CLSP of PRRSV | | Descriptor: | Non-structural protein, PHOSPHATE ION | | Authors: | Gao, F, Peng, H, Tian, X, Lu, G, Liu, Y, Gao, G.F. | | Deposit date: | 2008-11-17 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of Porcine Reproductive and Respiratory Syndrome Virus (PRRSV).
J.Mol.Biol., 392, 2009
|
|
7CMW
 
 | | Complex structure of PARP1 catalytic domain with pamiparib | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 1 | | Authors: | Feng, Y.C, Peng, H, Hong, Y, Liu, Y. | | Deposit date: | 2020-07-29 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of Pamiparib (BGB-290), a Potent and Selective Poly (ADP-ribose) Polymerase (PARP) Inhibitor in Clinical Development.
J.Med.Chem., 63, 2020
|
|
8D56
 
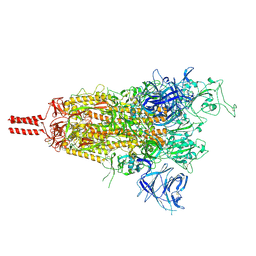 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D55
 
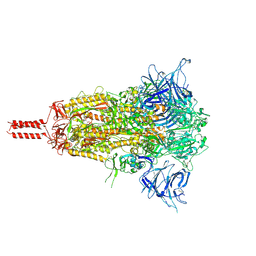 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D5A
 
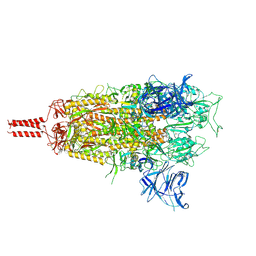 | | Middle state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6XR8
 
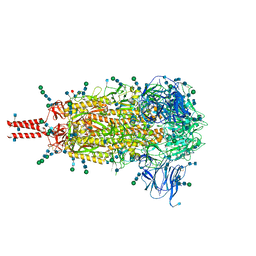 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
6XRA
 
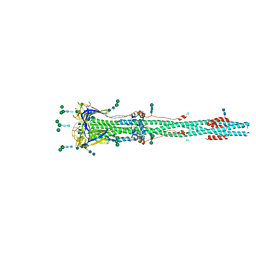 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Rits-Volloch, S, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
7SBR
 
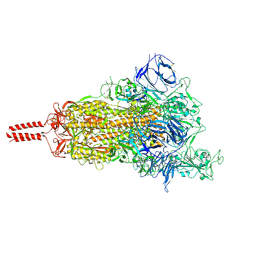 | | One RBD-up 2 of pre-fusion SARS-CoV-2 Kappa variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2021-09-25 | | Release date: | 2021-11-03 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant.
Science, 374, 2021
|
|
