4V4B
 
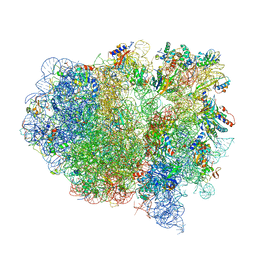 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
6AUI
 
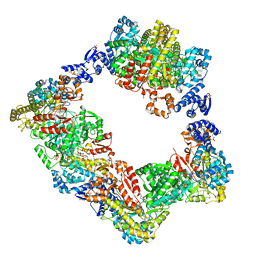 | | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Brignole, E.J, Drennan, C.L, Asturias, F.J, Tsai, K.L, Penczek, P.A. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 3.3- angstrom resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound.
Elife, 7, 2018
|
|
2NOQ
 
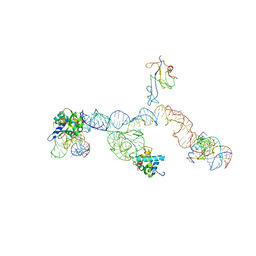 | | Structure of ribosome-bound cricket paralysis virus IRES RNA | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S5, ... | | Authors: | Schuler, M, Connell, S.R, Lescoute, A, Giesebrecht, J, Dabrowski, M, Schroeer, B, Mielke, T, Penczek, P.A, Westhof, E, Spahn, C.M.T. | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-21 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structure of the ribosome-bound cricket paralysis virus IRES RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4V5N
 
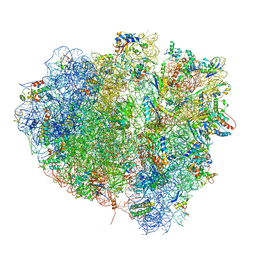 | | tRNA translocation on the 70S ribosome: the post- translocational translocation intermediate TI(POST) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-21 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
5AJ0
 
 | | Cryo electron microscopy of actively translating human polysomes (POST state). | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Behrmann, E, Loerke, J, Budkevich, T.V, Yamamoto, K, Schmidt, A, Penczek, P.A, Vos, M.R, Burger, J, Mielke, T, Scheerer, P, Spahn, C.M.T. | | Deposit date: | 2015-02-19 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Snapshots of Actively Translating Human Ribosomes
Cell(Cambridge,Mass.), 161, 2015
|
|
4V5M
 
 | | tRNA tranlocation on the 70S ribosome: the pre-translocational translocation intermediate TI(PRE) | | Descriptor: | 16S RRNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ratje, A.H, Loerke, J, Mikolajka, A, Bruenner, M, Hildebrand, P.W, Starosta, A.L, Doenhoefer, A, Connell, S.R, Fucini, P, Mielke, T, Whitford, P.C, Onuchic, J.N, Yu, Y, Sanbonmatsu, K.Y, Hartmann, R.K, Penczek, P.A, Wilson, D.N, Spahn, C.M.T. | | Deposit date: | 2010-10-01 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Head Swivel on the Ribosome Facilitates Translocation by Means of Intra-Subunit tRNA Hybrid Sites.
Nature, 468, 2010
|
|
6GY7
 
 | | Crystal structure of XaxB from Xenorhabdus nematophil | | Descriptor: | XaxB | | Authors: | Schubert, E, Raunser, S, Vetter, I.R, Prumbaum, D, Penczek, P.A. | | Deposit date: | 2018-06-28 | | Release date: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Membrane insertion of alpha-xenorhabdolysin in near-atomic detail.
Elife, 7, 2018
|
|
4A7F
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 3) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4A7N
 
 | | Structure of bare F-actin filaments obtained from the same sample as the Actin-Tropomyosin-Myosin Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, F-ACTIN | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4A7H
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 2) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
3J1Z
 
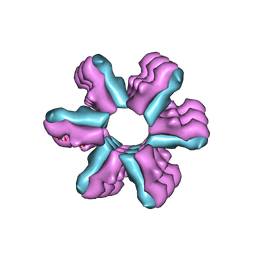 | | Inward-Facing Conformation of the Zinc Transporter YiiP revealed by Cryo-electron Microscopy | | Descriptor: | Cation efflux family protein | | Authors: | Coudray, N, Valvo, S, Hu, M, Lasala, R, Kim, C, Vink, M, Zhou, M, Provasi, D, Filizola, M, Tao, J, Fang, J, Penczek, P.A, Ubarretxena-Belandia, I, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2012-07-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6I3N
 
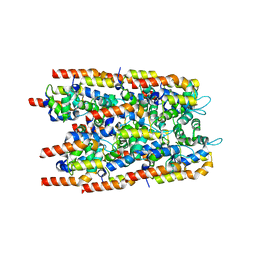 | |
6PU4
 
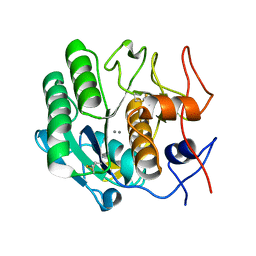 | | MicroED structure of proteinase K recorded on Falcon III | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Hattne, J, Martynowycz, M.W, Penzcek, P.A, Gonen, T. | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.1 Å) | | Cite: | MicroED with the Falcon III direct electron detector.
Iucrj, 6, 2019
|
|
6PU5
 
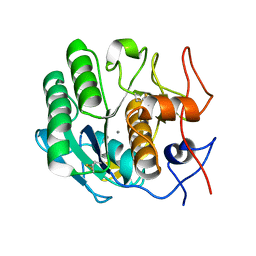 | | MicroED structure of proteinase K recorded on CetaD | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Hattne, J, Martynowycz, M.W, Penzcek, P.A, Gonen, T. | | Deposit date: | 2019-07-17 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.7 Å) | | Cite: | MicroED with the Falcon III direct electron detector.
Iucrj, 6, 2019
|
|
4P4H
 
 | |
3J8A
 
 | | Structure of the F-actin-tropomyosin complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | von der Ecken, J, Mueller, M, Lehman, W, Manstein, J.M, Penczek, A.P, Raunser, S. | | Deposit date: | 2014-10-08 | | Release date: | 2014-12-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the F-actin--tropomyosin complex.
Nature, 519, 2015
|
|
3J6J
 
 | | 3.6 Angstrom resolution MAVS filament generated from helical reconstruction | | Descriptor: | Mitochondrial antiviral-signaling protein | | Authors: | Wu, B, Peisley, A, Li, Z, Egelman, E, Walz, T, Penczek, P, Hur, S. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I.
Mol.Cell, 55, 2014
|
|
4V68
 
 | | T. thermophilus 70S ribosome in complex with mRNA, tRNAs and EF-Tu.GDP.kirromycin ternary complex, fitted to a 6.4 A Cryo-EM map. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schuette, J.-C, Spahn, C.M.T. | | Deposit date: | 2008-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | GTPase activation of elongation factor EF-Tu by the ribosome during decoding
Embo J., 28, 2009
|
|
6GY8
 
 | |
6GY6
 
 | |
5JQE
 
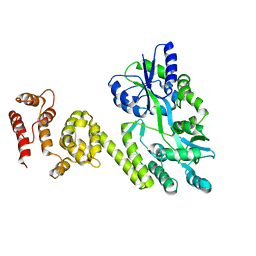 | | Crystal structure of caspase8 tDED | | Descriptor: | Sugar ABC transporter substrate-binding protein,Caspase-8 chimera | | Authors: | Fu, T, Li, Y, Lu, A, Wu, H. | | Deposit date: | 2016-05-04 | | Release date: | 2016-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.157 Å) | | Cite: | Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex.
Mol. Cell, 64, 2016
|
|
5L08
 
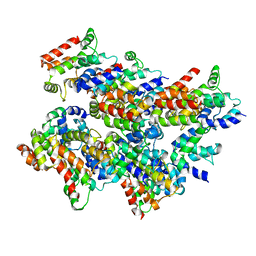 | | Cryo-EM structure of Casp-8 tDED filament | | Descriptor: | Caspase-8 | | Authors: | Fu, T.M, Li, Y, Lu, A, Wu, H. | | Deposit date: | 2016-07-26 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex.
Mol.Cell, 64, 2016
|
|
3J1O
 
 | |
3J1N
 
 | |
