5ON6
 
 | | Crystal structure of haemanthamine bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Meyer, M, Yusupova, G, Yusupov, M. | | Deposit date: | 2017-08-03 | | Release date: | 2018-02-28 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (3.10000229 Å) | | Cite: | The Amaryllidaceae Alkaloid Haemanthamine Binds the Eukaryotic Ribosome to Repress Cancer Cell Growth.
Structure, 26, 2018
|
|
6HHQ
 
 | | Crystal structure of compound C45 bound to the yeast 80S ribosome | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{S},4~{a}~{R},6~{S},7~{S},8~{a}~{R})-6,7-bis(chloranyl)-5,5,8~{a}-trimethyl-2-methylidene-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Pellegrino, S, Vanderwal, C.D, Yusupov, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-20 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (3.10000038 Å) | | Cite: | Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome.
Nucleic Acids Res., 47, 2019
|
|
4ABY
 
 | | Crystal structure of Deinococcus radiodurans RecN head domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4AD8
 
 | | Crystal structure of a deletion mutant of Deinococcus radiodurans RecN | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.998 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
4ABX
 
 | | Crystal structure of Deinococcus radiodurans RecN coiled-coil domain | | Descriptor: | DNA REPAIR PROTEIN RECN | | Authors: | Pellegrino, S, Radzimanowski, J, de Sanctis, D, McSweeney, S, Timmins, J. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-12 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | Structural and Functional Characterization of an Smc-Like Protein Recn: New Insights Into Double-Strand Break Repair.
Structure, 20, 2012
|
|
6GQ1
 
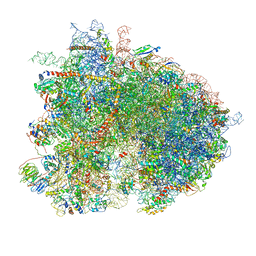 | | Cryo-EM reconstruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GMPPCP/sordarin) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pellegrino, S, Demeshkina, N, Mancera-Martinez, E, Melnikov, S, Simonetti, A, Myasnikov, A, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-11 | | Last modified: | 2018-08-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
7R4X
 
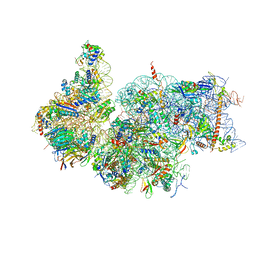 | | Cryo-EM reconstruction of the human 40S ribosomal subunit - Full map | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Pellegrino, S, Dent, K.C, Spikes, T, Warren, A.J. | | Deposit date: | 2022-02-09 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Cryo-EM reconstruction of the human 40S ribosomal subunit at 2.15 angstrom resolution.
Nucleic Acids Res., 51, 2023
|
|
6GQB
 
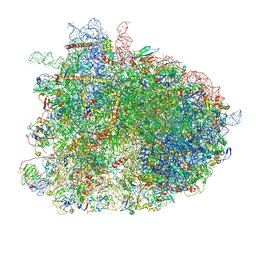 | | Cryo-EM reconstruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GDP+AlF4/sordarin) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pellegrino, S, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-11 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
6GQV
 
 | | Cryo-EM recosntruction of yeast 80S ribosome in complex with mRNA, tRNA and eEF2 (GMPPCP) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Pellegrino, S, Yusupov, M, Yusupova, G, Hashem, Y. | | Deposit date: | 2018-06-08 | | Release date: | 2018-07-11 | | Last modified: | 2018-08-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Insights into the Role of Diphthamide on Elongation Factor 2 in mRNA Reading-Frame Maintenance.
J. Mol. Biol., 430, 2018
|
|
5TBW
 
 | | Crystal structure of chlorolissoclimide bound to the yeast 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Konst, Z.A, Szklarski, A.R, Pellegrino, S, Michalak, S.E, Meyer, M, Zanette, C, Cencic, R, Nam, S, Horne, D.A, Pelletier, J, Mobley, D.L, Yusupova, G, Yusupov, M, Vanderwal, C.D. | | Deposit date: | 2016-09-13 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis facilitates an understanding of the structural basis for translation inhibition by the lissoclimides.
Nat Chem, 9, 2017
|
|
5MEI
 
 | | Crystal structure of Agelastatin A bound to the 80S ribosome | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | McClary, B, Zinshteyn, B, Meyer, M, Jouanneau, M, Pellegrino, S, Yusupova, G, Schuller, A, Reyes, J.C.P, Lu, J, Luo, C, Dang, Y, Romo, D, Yusupov, M, Green, R, Liu, J.O. | | Deposit date: | 2016-11-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of Eukaryotic Translation by the Antitumor Natural Product Agelastatin A.
Cell Chem Biol, 24, 2017
|
|
7QH6
 
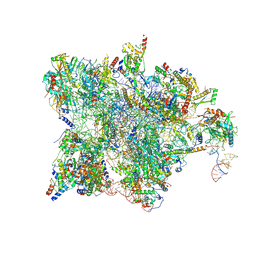 | | Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 1 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L13, mitochondrial, ... | | Authors: | Rebelo-Guiomar, P, Pellegrino, S, Dent, K.C, Warren, A.J, Minczuk, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit.
Nat Commun, 13, 2022
|
|
7QH7
 
 | | Cryo-EM structure of the human mtLSU assembly intermediate upon MRM2 depletion - class 4 | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Rebelo-Guiomar, P, Pellegrino, S, Dent, K.C, Warren, A.J, Minczuk, M. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | A late-stage assembly checkpoint of the human mitochondrial ribosome large subunit.
Nat Commun, 13, 2022
|
|
6SJ6
 
 | | Cryo-EM structure of 50S-RsfS complex from Staphylococcus aureus | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Khusainov, I, Pellegrino, S, Yusupova, G, Yusupov, M, Fatkhullin, B. | | Deposit date: | 2019-08-12 | | Release date: | 2020-04-08 | | Last modified: | 2020-07-08 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach.
Nat Commun, 11, 2020
|
|
8A3D
 
 | | Human mature large subunit of the ribosome with eIF6 and homoharringtonine bound | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 28S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Faille, A, Warren, A.J, Dent, K.C. | | Deposit date: | 2022-06-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | The chemical landscape of the human ribosome at 1.67 angstrom resolution.
Biorxiv, 2023
|
|
4L3C
 
 | | Structure of HLA-A2 in complex with D76N b2m mutant and NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-05 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
4L29
 
 | | Structure of wtMHC class I with NY-ESO1 double mutant | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, GLYCEROL, ... | | Authors: | Halabelian, L, Giorgetti, S, Bellotti, V, Bolognesi, M, Ricagno, S. | | Deposit date: | 2013-06-04 | | Release date: | 2013-12-25 | | Last modified: | 2014-02-26 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Class I Major Histocompatibility Complex, the Trojan Horse for Secretion of Amyloidogenic beta 2-Microglobulin.
J.Biol.Chem., 289, 2014
|
|
5JWE
 
 | | Crystal structure of H-2Db in complex with the LCMV-derived GP92-101 peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Buratto, J, Badia-Martinez, D, Norstrom, M, Sandalova, T, Achour, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of H-2Db in complex with the LCMV-derived peptides GP92 and GP392 explain pleiotropic effects of glycosylation on antigen presentation and immunogenicity.
PLoS ONE, 12, 2017
|
|
7P0A
 
 | |
7P0T
 
 | |
4OJH
 
 | |
4OJ3
 
 | |
4OJG
 
 | |
6SJ5
 
 | | Crystal structure of the uL14-RsfS complex from Staphylococcus aureus | | Descriptor: | 50S ribosomal protein L14, ACETIC ACID, PHOSPHATE ION, ... | | Authors: | Fatkhullin, B, Gabdulkhakov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2019-08-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.26686549 Å) | | Cite: | Mechanism of ribosome shutdown by RsfS in Staphylococcus aureus revealed by integrative structural biology approach.
Nat Commun, 11, 2020
|
|
6GB5
 
 | | Structure of H-2Db with truncated SEV peptide and GL | | Descriptor: | Beta-2-microglobulin, GLY-LEU, GLYCEROL, ... | | Authors: | Hafstrand, I, Sandalova, T, Achour, A. | | Deposit date: | 2018-04-13 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Successive crystal structure snapshots suggest the basis for MHC class I peptide loading and editing by tapasin.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
