3HQJ
 
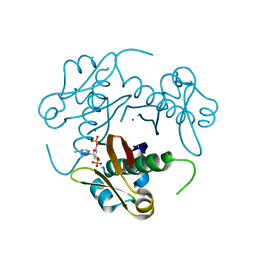 | | Structure-function analysis of Mycobacterium tuberculosis acyl carrier protein synthase (AcpS). | | Descriptor: | COENZYME A, Holo-[acyl-carrier-protein] synthase, MAGNESIUM ION | | Authors: | Dym, O, Albeck, S, Peleg, Y, Schwarz, A, Shakked, Z, Burstein, Y, Zimhony, O, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2009-06-07 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-function analysis of the acyl carrier protein synthase (AcpS) from Mycobacterium tuberculosis.
J.Mol.Biol., 393, 2009
|
|
6HAP
 
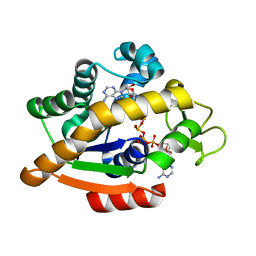 | | Adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
7O4D
 
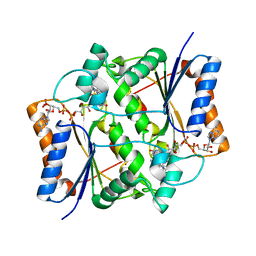 | | QR2 inhibitor from a novel sulfanamide series to tackle age related oxidative stress and cognitive decline | | Descriptor: | 8-methyl-2-(4-methyl-3-piperazin-1-ylsulfonyl-phenyl)imidazo[1,2-a]pyridine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Gould, N.L, Scherer, G.R, Carvalh, S, Shurrush, K, Edry, E, Elkobi, A, David, O, Dym, O, Albeck, S, Peleg, Y, Germain, N, Babaev, I, Sharir, H, Lefker, B, Subramanyam, C, Barr, H, Rosenblum, K. | | Deposit date: | 2021-04-06 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Specific quinone reductase 2 inhibitors reduce metabolic burden and reverse Alzheimer's disease phenotype in mice.
J.Clin.Invest., 133, 2023
|
|
6HAM
 
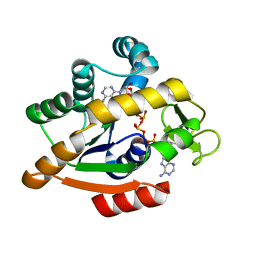 | | Adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
2V9D
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-23 | | Release date: | 2008-03-04 | | Last modified: | 2019-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
2V8Z
 
 | | Crystal Structure of YagE, a prophage protein belonging to the dihydrodipicolinic acid synthase family from E. coli K12 | | Descriptor: | YAGE | | Authors: | Manicka, S, Peleg, Y, Unger, T, Albeck, S, Dym, O, Greenblatt, H.M, Bourenkov, G, Lamzin, V, Krishnaswamy, S, Sussman, J.L. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Yage, a Putative Dhdps Like Protein from Escherichia Coli K12.
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
7PL9
 
 | |
3D3M
 
 | |
6QJZ
 
 | |
5HPZ
 
 | | type II water soluble Chl binding proteins | | Descriptor: | 13'2-hydroxyl-Chlorophyll a, Water-soluble chlorophyll protein | | Authors: | Bednarczyk, D, Dym, O, Prabahard, V, Noy, D. | | Deposit date: | 2016-01-21 | | Release date: | 2016-05-04 | | Last modified: | 2019-05-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Fine Tuning of Chlorophyll Spectra by Protein-Induced Ring Deformation.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6YBH
 
 | | Deoxyribonucleoside Kinase | | Descriptor: | Deoxynucleoside kinase, GLYCEROL, Pyrrolo-dC, ... | | Authors: | Allouche-Arnon, H, Bar-Shir, A, Dym, O. | | Deposit date: | 2020-03-17 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computationally designed dual-color MRI reporters for noninvasive imaging of transgene expression.
Nat.Biotechnol., 40, 2022
|
|
6ZBS
 
 | | Beta ODAP Synthetase (BOS) | | Descriptor: | Beta ODAP Synthetase (BOS), DI(HYDROXYETHYL)ETHER, MAGNESIUM ION | | Authors: | Dym, O. | | Deposit date: | 2020-06-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification and characterization of the key enzyme in the biosynthesis of the neurotoxin beta-ODAP in grass pea.
J.Biol.Chem., 298, 2022
|
|
6GG1
 
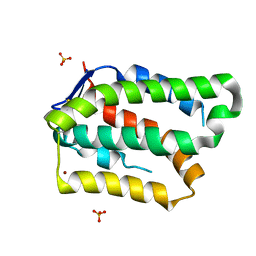 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
6GJC
 
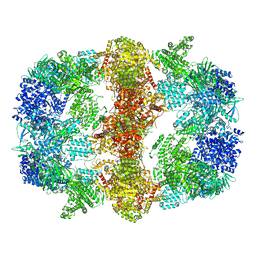 | | Structure of Mycobacterium tuberculosis Fatty Acid Synthase - I | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase | | Authors: | Elad, N, Baron, S, Shakked, Z, Zimhony, O, Diskin, R. | | Deposit date: | 2018-05-16 | | Release date: | 2018-09-05 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of Type-I Mycobacterium tuberculosis fatty acid synthase at 3.3 angstrom resolution.
Nat Commun, 9, 2018
|
|
7AUE
 
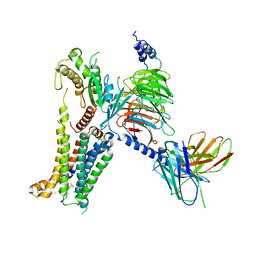 | | Melanocortin receptor 4 (MC4R) Gs protein complex | | Descriptor: | AMINOSERINE, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Degtjarik, O, Israeli, H, Prabahar, V, Shalev-Benami, M. | | Deposit date: | 2020-11-02 | | Release date: | 2021-04-28 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure reveals the activation mechanism of the MC4 receptor to initiate satiation signaling.
Science, 372, 2021
|
|
4PTN
 
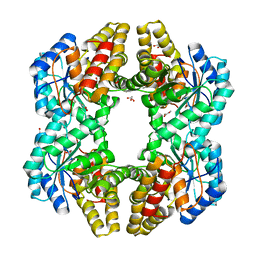 | | Crystal Structure of YagE, a KDG aldolase protein in complex with Magnesium cation coordinated L-glyceraldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, L-glyceraldehyde, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-03-11 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a putative DHDPS-like protein from Escherichia coli K12.
Proteins, 71, 2008
|
|
3TPP
 
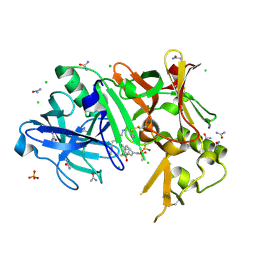 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPJ
 
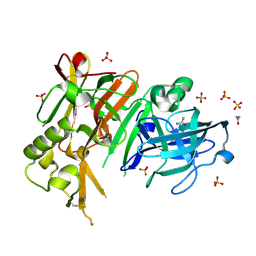 | | APO structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPR
 
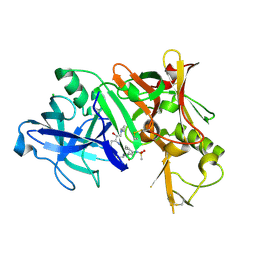 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPL
 
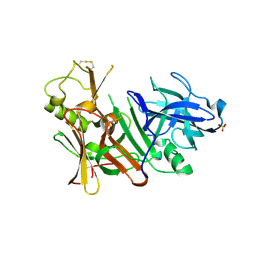 | | APO Structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4OE7
 
 | | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal | | Descriptor: | (4R)-4-hydroxy-2,5-dioxopentanoic acid, (4S)-4-hydroxy-2,5-dioxopentanoic acid, 1,2-ETHANEDIOL, ... | | Authors: | Manoj Kumar, P, Baskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-12 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein, in complex with aldol condensed product of pyruvate and glyoxal
To be Published
|
|
4ONV
 
 | | Crystal structure of YagE, a KDG aldolase protein in complex with 2-Keto-3-deoxy gluconate | | Descriptor: | 1,2-ETHANEDIOL, 2-KETO-3-DEOXYGLUCONATE, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Bhaskar, V, Manicka, S, Krishnaswamy, S. | | Deposit date: | 2014-01-29 | | Release date: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structure of YagE, a KDG aldolase protein in complex with 2-Keto-3-deoxy gluconate
To be Published
|
|
